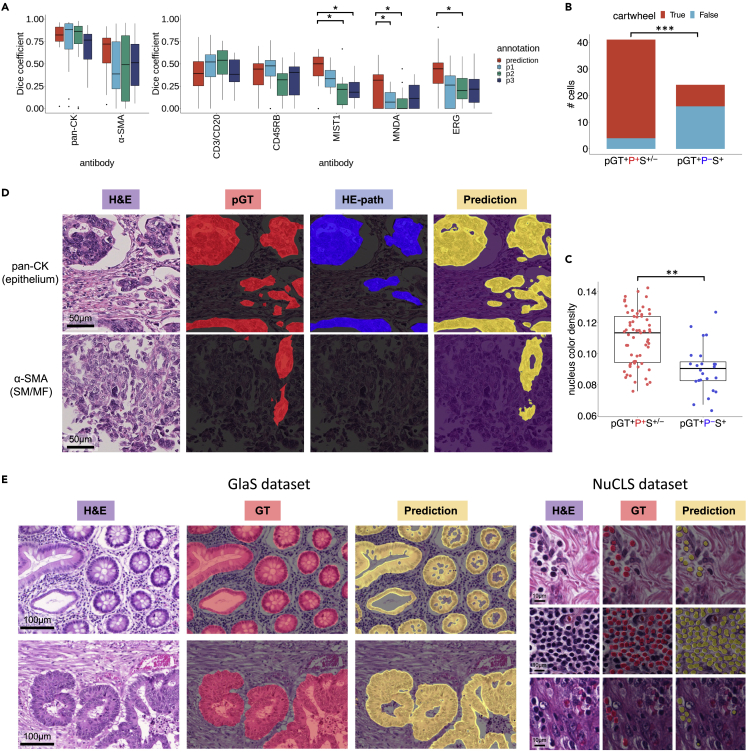
Figure 6.
Performance evaluation of the segmentation models trained on the generated annotation masks
(A) Comparison of the annotation accuracies between pathologists and prediction of the segmentation models in terms of the Dice coefficient (F1 score) (n = 10 patches of 217.5 × 217.5 μm for each tissue or cell type). The optimal segmentation model in terms of validation loss was applied, and each point in the box plots represents a patch. pGT annotations were made by pathologists who evaluated both H&E and the corresponding IF images. Regions or cells annotated by at least two of the three pathologists were used. p < 0.05 was considered statistically significant, as shown by asterisks. See also Figure S9 and Table S5.
(B) Distribution of plasma cells with or without typical cartwheel-shaped nuclei (n = 41 cells for pGT+P+S+/− and n = 27 cells for pGT+P−S+).
(C) Nuclear hematoxylin intensity of lymphocytes (n = 63 cells for pGT+P+S+/− and n = 24 cells for pGT+P−S+). For the box plot, the lower and upper hinges correspond to the 25th and 75th percentiles, respectively; the upper whisker extends from the hinge to the largest value no further than 1.5× IQR from the hinge. The lower whisker extends from the hinge to the smallest value at 1.5× IQR of the hinge. pGT, ground truth; P, HE-path; S, prediction by the segmentation model. ∗∗∗p < 0.0001, ∗∗p < 0.01. See also Figure S10.
(D) Comparison of pathologist annotations for AE1/3 and SMA.
(E) Samples of the segmentation results based on GlaS and NuCLS for epithelium and lymphocytes, respectively. Note that the segmentation models were trained using only the SegPath dataset, and no fine-tuning was performed on the target dataset. See also Figures S11–S15.