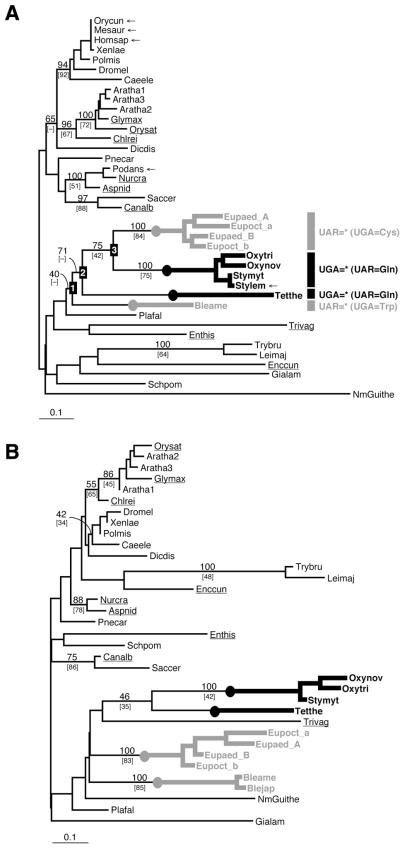
Figure 1.
(A) An unrooted eRF1 tree based on the FL dataset using the Γ-FITCH protein distance method. Abbreviations of the sequence names are listed in Table 2. BP are listed for major nodes. Percent occurrence in 10 000 quartet puzzling trees are given in brackets. Dashes indicate that given nodes were not supported by bootstrap or quartet puzzling analyses. The novel sequences determined or identified in this study are underlined. The redundant sequences removed from the analyses using the D1 dataset are marked by arrows. The ciliate lineages that independently converted the standard code to variant codes [UGA = * (UAR = Gln) and UAR = * (UGA = Cys or Trp)] are highlighted by thick black and gray lines, respectively. (B) An unrooted eRF1 tree based on the D1 dataset using the Γ-FITCH protein distance method.