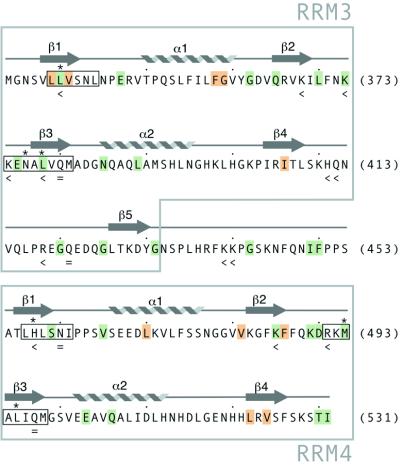
Figure 3.
Overview of amino acids residues in PTB-34 with significant chemical shift changes upon binding of RNA. Residues with a combined shift of perturbation of ≥0.1 p.p.m. for at least four out of the six RNA oligonucleotides which bound to the protein are shaded green or orange. Green shading indicates that the amino acid side-chain is solvent-exposed; buried side-chains are shaded orange. The hexameric RNP-2 and octameric RNP-1 motifs in both RRMs are boxed. Asterisks mark the positions of residues that are aromatic in most RRM sequences (48). Mutation of residues marked with ‘<’ reduced the RNA binding affinity; residues marked with ‘=’ had no effect on RNA binding affinity (29).