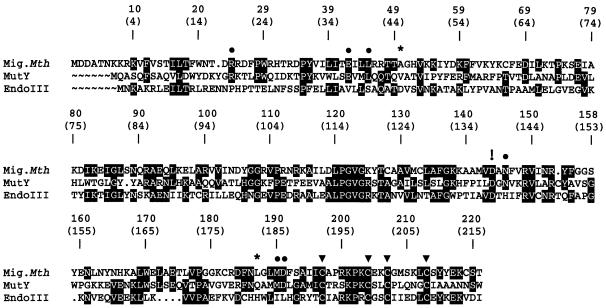
Figure 1.
Amino acid alignment of Mig.MthI (NC_001336) (4), MutY.Eco (M59471) (10) and Endonuclease III (J02857) (12); GenBank accession numbers are given in parentheses. The ClustalW algorithm (24) was used for alignment. The C-terminal 134 residues of MutY are not represented. MutY residues that contact the mismatched adenine base (11) and which are conserved in Mig.MthI are indicated by filled circles. Asterisks indicate MutY amino acid residues involved in adenine binding that are not conserved in Mig.MthI. The four cystein residues contributing the sulfur part to the [4Fe-4S] cluster are highlighted by filled triangles. The exclamation mark points out the aspartic acid residue involved in acid/base catalysis (11). Shading highlights sequence positions with amino acid residues conserved between at least two enzymes.