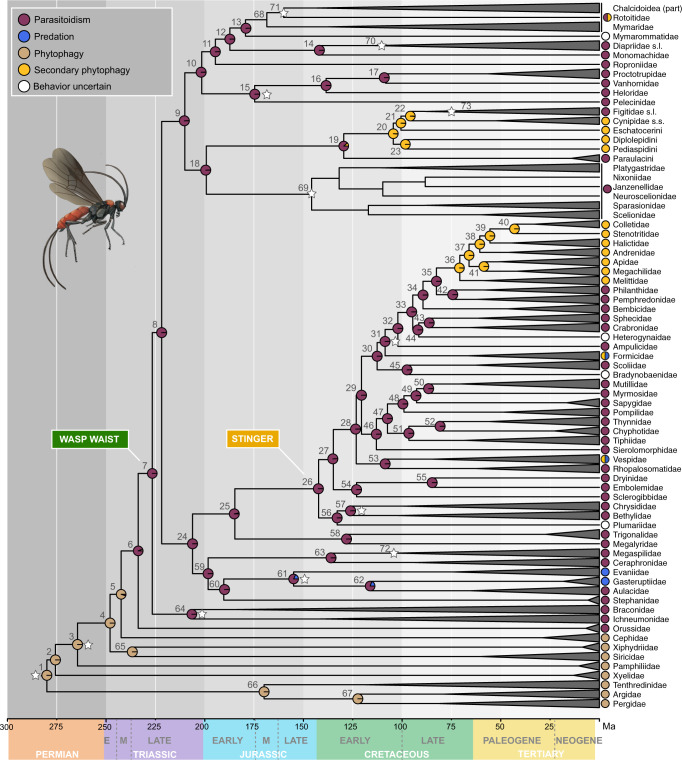
Fig. 2. Timeline and evolution of parasitoidism in Hymenoptera.
Chronogram estimated using approximate likelihood in MCMCTREE with the PAMLv4.9 package113 from the nuc-70% matrix and topology C-1. All outgroups were pruned from the tree and alignment prior to divergence time estimation. Twelve calibration points were used, which are indicated by a white star. Terminals have been collapsed down to family or clade-level post analysis. Families of Platygastroidea are shown in the chronogram, but were lumped at the superfamily level for ancestral state reconstructions; therefore, pies are absent in this clade. Ancestral state reconstructions with corHMM were estimated from topology C-1 and mapped in pie format onto the chronogram; states for terminals are indicated beside terminal branches. Divergence estimates and ancestral state probabilities can be accessed in Supplementary Data 6 and 8, referring to numbers beside nodes. Pie states are red = parasitoid, blue = predatory, brown = phytophagous, yellow = secondarily phytophagous, and white = behavior unknown. For detailed information on methodology and results, also refer to the main text and the Supplementary Methods. Source data for this figure can be found in the Dryad repository at 10.5061/dryad.08kprr54m (folder 2.2.2).