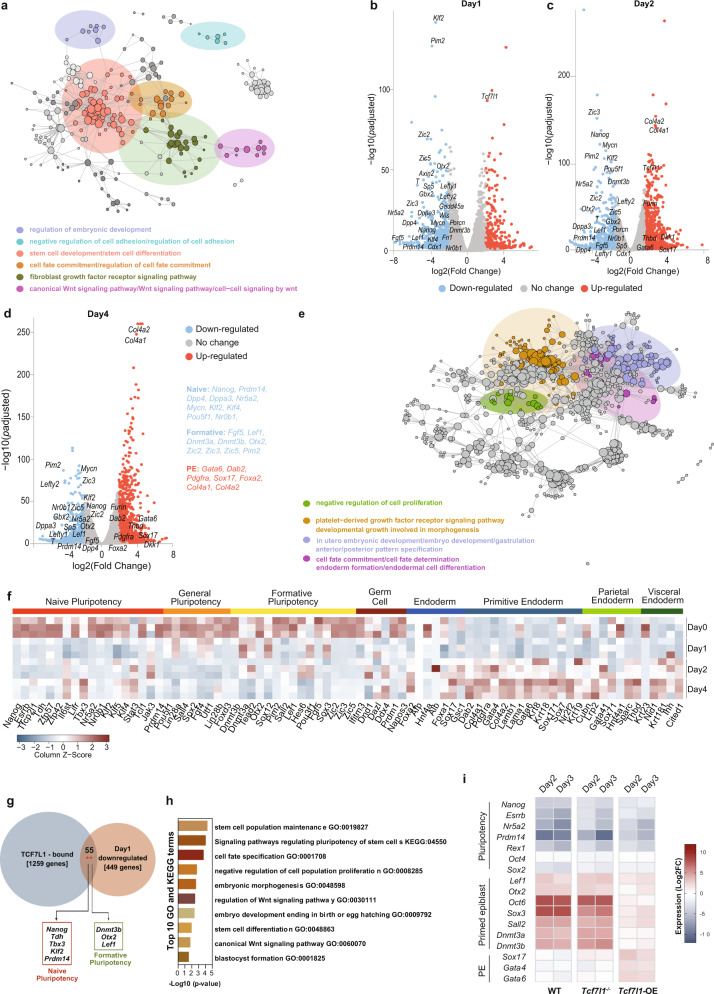
Fig. 5. PE cell fate specification is preceded by naive and formative pluripotency repression by TCF7L1.
a GO and KEGG functional enrichment analysis for Tcf7l1-OE-mESCs after 24 H of induction. Functional term networks were obtained using Cytoscape’s ClueGO plugin. Each node corresponds to an enriched term and edges connect terms that share a significant number of genes. Network communities (shown in color) were created using the Louvain algorithm, adjusted for betweenness centrality. b–d Volcano plots showing down- (blue dots) and upregulated (red dots) genes altered on different timepoints of Tcf7l1 induction. Gene values are reported as a Log2FoldChange. Differential expression analysis was performed with a two-tailed Wald’s test. All p-values were adjusted for multiple comparisons using Benjamini-Hochberg correction. Annotated points correspond to selected marker genes. b DEGs after 24H of induction. c DEGs after 2 days of induction. d DEGs after 4 days of induction. e GO and KEGG functional enrichment analysis for Tcf7l1 mESCs after 4 days of induction. Network was created and analyzed as described in Fig. 5a. f Normalized gene counts heatmap showing 80 selected specific lineage markers in WT and Tcf7l1−/− cells upon 1, 2 and 4 days of induction. g Venn diagram indicating genes bound by TCF7L1 (1259 genes) from publicly available ChIP-seq data36 and downregulated genes in Tcf7l1-OE-mESCs after 1 day of induction (449 genes with unique promoters). There were 55 genes shared between the two sets (p-value of a two-tailed Fisher’s exact test = 0.0021) which include markers of naive and formative pluripotency. h Top 10 functional enrichment terms for the 55 genes downregulated in Tcf7l1-OE-mESCs after 1 day of induction and bound by TCF7L1. i qRT-PCR of pluripotency, primed epiblast/formative, and PE gene markers in WT, Tcf7l1−/− and Tcf7l1-OE-mESCs upon EpiLCs differentiation. Gene expression values are reported as Log2 of fold change expression, see Source data file.