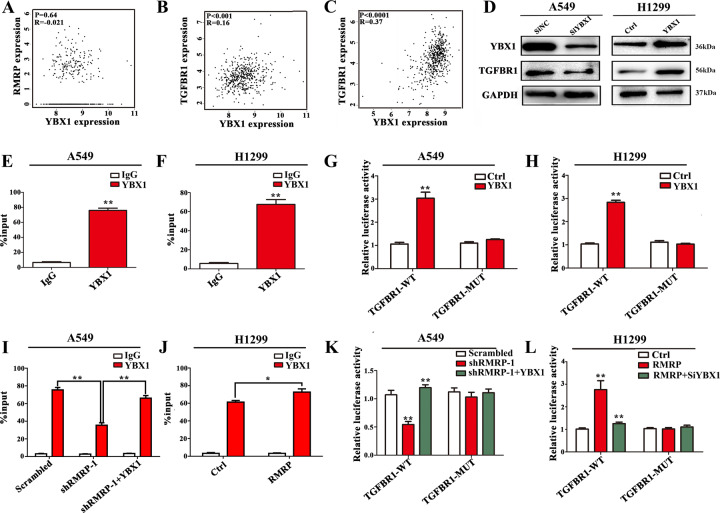
Fig. 6. RMRP promotes TGFBR1 transcription by recruiting YBX1 to the TGFBR1 promoter.
A The expression of YBX1 was not correlated with RMRP in TCGA-LUAD database by GEPIA (http://gepia.cancer-pku.cn/) using Pearson Correlation Coefficient. B The expression of YBX1 showed a positive correlation with TGFBR1 in TCGA-Lung adenocarcinoma (LUAD) database by GEPIA using Pearson Correlation Coefficient. C The expression of YBX1 showed a positive correlation with TGFBR1 in TCGA-Kidney Renal Clear Cell Carcinoma (KIRC) database by GEPIA using Pearson Correlation Coefficient. D Results of Western blot assays: knockdown of YBX1 inhibited the expression of TGFBR1, while overexpression of YBX1 increased the expression of TGFBR1 (n = 3). E, F YBX1-bound complex showed remarkable enrichment of the TGFBR1 promoter by ChIP and quantitative RT-PCR analysis in A549 and H1299 cells. IgG was used as a negative control. Enrichment was quantified relative to input controls (n = 3). G, H Luciferase activity assays were performed in A549 and H1299 cells transfected with wide type (TGFBR1-WT) or mutant-type (TGFBR1-MUT) TGFBR1 promoter-containing pGL3 reporter vector. These cells were further transfected with YBX1 (YBX1) or control (Ctrl). After 48 h, firefly luciferase activity was detected and normalized to Renilla luciferase activity (n = 3). I, J ChIP and quantitative RT-PCR analysis in the treated A549 and H1299 cells. IgG was used as a negative control. Enrichment was quantified relative to input controls (n = 3). K, L Luciferase activity assays in the treated A549 and H1299 cells were further transfected with TGFBR1-MUT or TGFBR1-WT. After 48 h, firefly luciferase activity was detected and normalized to Renilla luciferase activity, respectively (n = 3). Data presented as mean ± SD from three independent experiments. Student’s t test was used for statistical analysis: *P < 0.05; **P < 0.01; ***P < 0.001.