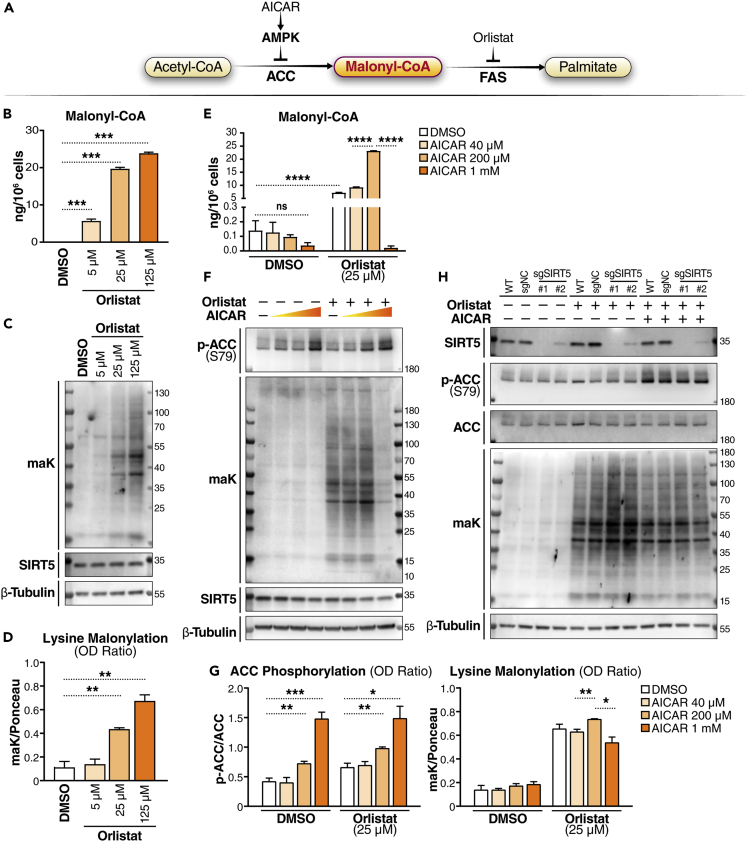
Figure 1.
Malonyl-CoA availability affects lysine malonylation
(A) Schematic of the fatty acid synthesis pathway and compounds affecting malonyl-CoA level. AMPK, AMP-activated protein kinase; ACC, acetyl-CoA carboxylase; FAS, fatty acid synthase.
(B) Malonyl-CoA levels in K562 cells treated with orlistat at different doses as indicated for 24 h were measured with LC-MS. [13C3]-malonyl-CoA was added to each sample as a standard for quantification. Quantified levels are normalized to cell count in each sample. N = 3 per treatment. Values are shown as mean ± SEM. ∗∗∗p < 0.001 using unpaired Student’s t test.
(C) Malonyl-CoA levels in K562 cells treated with orlistat and AICAR at different doses as indicated for 24 h were measured with LC-MS. [13C3]-malonyl-CoA was added to each sample as a standard for quantification. Quantified levels are normalized to cell count in each sample. N = 3 per treatment. Values are shown as mean ± SEM. ns (not significant) ≧ 0.05, and ∗∗∗∗p < 10−4 using unpaired Student’s t test.
(D) Western blot (WB) of lysine malonylation (maK) in K562 cells at 24 h after orlistat treatment at the indicated concentrations.
(E) Quantification of WBs by calculating the OD ratio of maK to Ponceau S staining. N = 3. Values are shown as mean ± SEM. ∗∗p < 0.01 using unpaired Student’s t test.
(F) WB of maK in K562 cells at 24 h after drug treatment. Orlistat: 25 μM; AICAR: 40 μM, 200 μM, or 1 mM.
(G) Quantification of WBs by calculating the OD ratio of phosphorylated ACC (at Serine 79) to total ACC, and OD ratio of maK to Ponceau S staining. N = 3. Values are shown as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 using unpaired Student’s t test.
(H) WB of maK in K562 (wildtype) cells, and K562-dCas9-KRAB cells expressing sgNC, sgSIRT5#1 or sgSIRT5#2 at 24 h after drug treatment. Orlistat: 25 μM; AICAR: 1 mM.