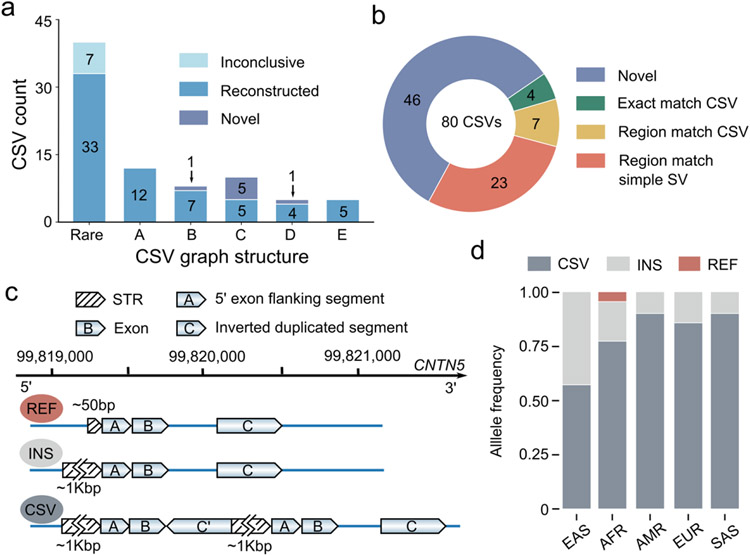
Fig. 2 ∣. Application of Svision on HG00733 HiFi data.
a, The Svision complex structural variants (CSVs) overlapping with Phased Assembly Variant (PAV) calls and reconstructed with phased HiFi contigs. The rare type represented multiple CSV graph structures, each of which contains fewer than five complex events. A: Inverted duplication. B: Deletion associated with inversion. C: Deletion associated with inverted duplication. D: Multiple deletion with spacer. E: Deletion associated with duplication. Inconclusive: Unable to characterize visually. Reconstructed: SVision CSV structure validated via contig based manual curation. Novel: No overlapping PAV call. b, Compared to 80 CSVs in HG00733 detected by SVision, short-read callset correctly reported the full structure of 4 CSVs (exact match), partially called 7 (region match), misinterpreted 23 events as simple (region match) and completely missed 46 CSVs. c, Three distinct alleles were found for the CNTN5 locus, including a contracted tandem repeat (REF), an expanded tandem repeat (INS), and an expanded tandem repeat with a complex duplication containing CNTN5 exon 4. d, The frequencies of three CNTN5 alleles differ among populations.