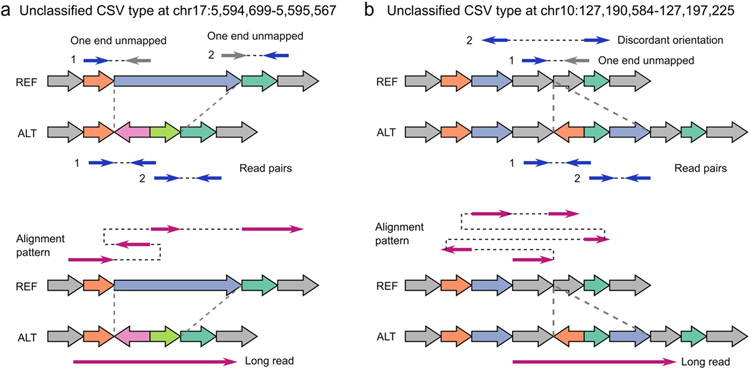
Extended Data Fig. 4 ∣. The diagrams and alignment patterns of two unclassified complex structural variants.
a, SVision correctly detected a deleted sequence replaced with dispersed duplication and inverted duplication. b, SVision characterized a complex insertion, consisting of two dispersed duplications and one inverted duplication. Both types of (a) and (b) are labeled as unclassified (NA) in the 1KGP call set. The top panel of (a) and (b) are the discordant alignments derived from short-read sequencing (i.e., one end unmapped and discordant alignment). The bottom panels of (a) and (b) describe the abnormal alignments from long-read alignment.