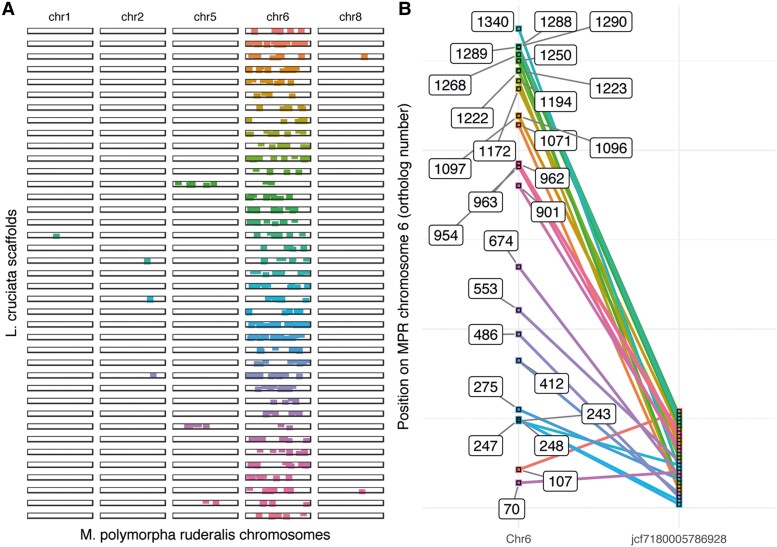
Fig. 4.
Chromosomal inversions can explain much of the decrease in collinearity between M. polymorpha and L. cruciata. (A) Illustration of rearrangements within and among chromosomes, exemplified by the 36 largest L. cruciata scaffolds with at least 10 orthologous hits on M. polymorpha subsp. ruderalis chromosome 6. Each squared dot represents one gene and all single-copy orthologous genes on the L. cruciata scaffolds are printed including the few genes having their orthologous hit on another chromosome than chromosome 6. The scaffolds are color-coded. (B) A close-up of the largest L. cruciata scaffold (in the number of orthologous single-copy genes) and its collinear relationship with M. polymorpha subsp. ruderalis chromosome 6. The genes are numbered after their order of appearance on chromosome 6—gaps in the numbering result from M. polymorpha genes that do not have an identified ortholog on this L. cruciata scaffold. The lines connecting orthologs have different colors for the sake of clarity.