Table 3.
Features that were confirmed using MAW and the original study on the bryophytes data
| Precursor mass [m/z] | Retention time [s] | Molecular formula | Molecular structure | Chemical class | Name of the compound | MSI-level | Rank |
|---|---|---|---|---|---|---|---|
| 425.176 | 607.49 | C28H26O4 | 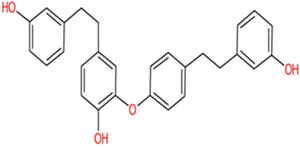 |
Lignans, neolignans and related compounds | Perrottetin E | 3 | 10 |
| 182.081 | 47.16 | C9H11NO3 | 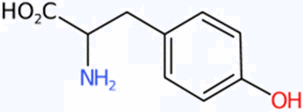 |
Carboxylic acids and derivatives | Tyrosine | 2 | 2 |
| 455.155 | 220.91 | C28H24O6 | 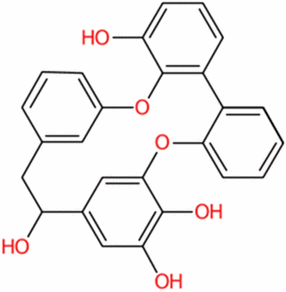 |
Lignans, neolignans and related compounds | Marchantin D | 3 | 23 |
| 287.056 | 318.26 | C15H10O6 |  |
Flavonoids | Kaempferol | 2 | 8 |
| 165.055 | 484.08 | C9H8O3 | 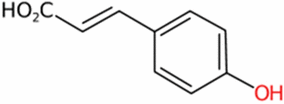 |
Cinnamic acids and derivatives | 4-Hydroxycinnamic acid | 2 | 6 |
| 291.231 | 773.87 | C18H32O2 | 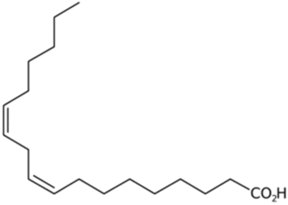 |
Fatty acyls | Linoleate | 3 | 6 |
| 379.283 | 702.51 | C21H38O4 |  |
Fatty acyls | Glyceryl monolinoleate | 3 | 4 |
| 255.231 | 859.92 | C16H32O2 | Fatty acyls | Palmitate | 3 | 1 |
The precursor mass [m/z] and retention time [s] represent the mass of the precursor ions given in mass-to-charge ratio and the median retention time of the precursor ion given in seconds, respectively. The molecular formula and name of the compound are extracted using PubChemPy or SIRIUS, depending on the annotation source. The chemical classes of the compounds are either extracted from ClassyFire or CANOPUS. All the molecular structures are generated via CDK-Depict. The MSI-levels are in accordance with the rules described in Table 1. The given ranks are from MAW, using the database COCONUT in the compound dereplication module
