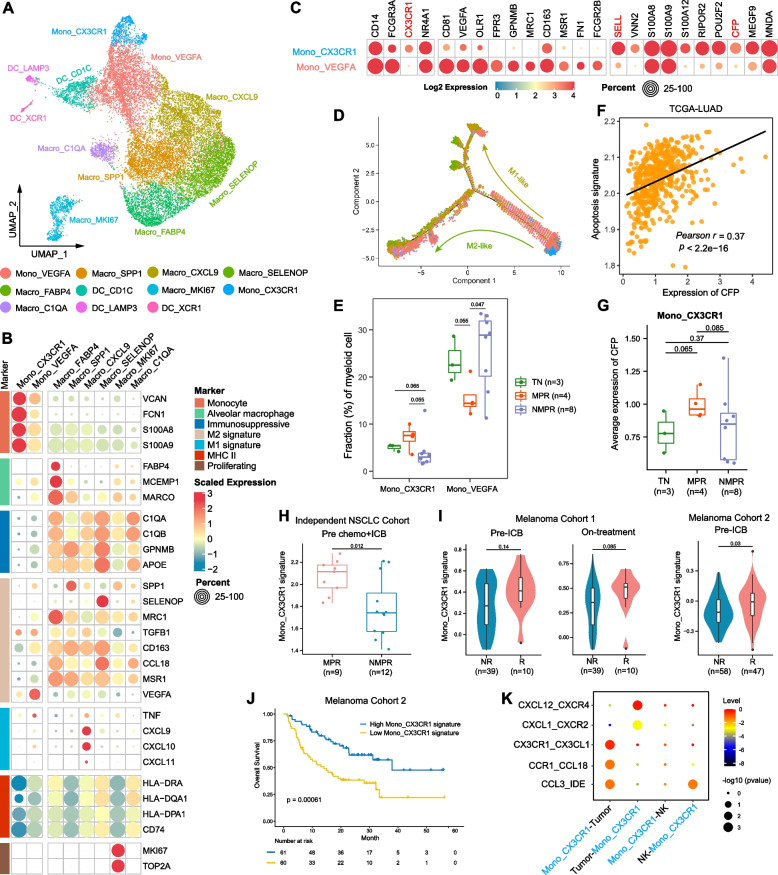
Fig. 5.
Monocyte remodeling after therapy. A UMAP plot of myeloid cells colored by clusters. B Heatmap of normalized expression of monocyte/macrophage marker genes among clusters. C Heatmap of selected marker genes of defined monocyte clusters. D The developmental trajectory of monocyte/macrophages inferred by Monocle2. The Mono_CX3CR1 cells were the roots of trajectory, and differentiated into M1-like (Macro_CXCL9) or M2-like (Macro_SELENOP and Macro_C1QA) cells. E Boxplot showing cellular fractions of each monocyte cluster in TN (n = 3), MPR (n = 4), and NMPR (n = 8) patients. Center line indicates the median, lower, and upper hinges represent the 25th and 75th percentiles, respectively, and whiskers denote 1.5× interquartile range. All differences with P < 0.10 are indicated. One-sided unpaired Wilcoxon test was used. F Scatter diagram showing a significantly positive correlation between expression level of CFP and apoptosis signature in TCGA-LUAD patients. P values were determined by two-sided Pearson correlation test. G Boxplots of the average expression of CFP in Mono_CX3CR1 cells in TN (n = 3), MPR (n = 4), and NMPR (n = 8) patients. One-sided t-test was used. H Violin and box plots of Mono_CX3CR1 signature in our validation cohort (9 patients were assessed as MPR and 12 as NMPR after surgery) before ICB + chemotherapy. One-sided unpaired Wilcoxon test was used. I Violin and box plots of Mono_CX3CR1 signature in responders (R) and non-responders (NR, removing SD patients) in advanced melanoma cohorts. Two-sided unpaired Wilcoxon test was used. J Kaplan–Meier survival curve of the signature of Mono_CX3CR1 in melanoma cohort 2. Survival curves were compared by the Log-Rank test. K Summary of selected ligand-receptor interactions from CellPhoneDB among Mono_CX3CR1 cells, cancer cells, and NK cells in MPR patients