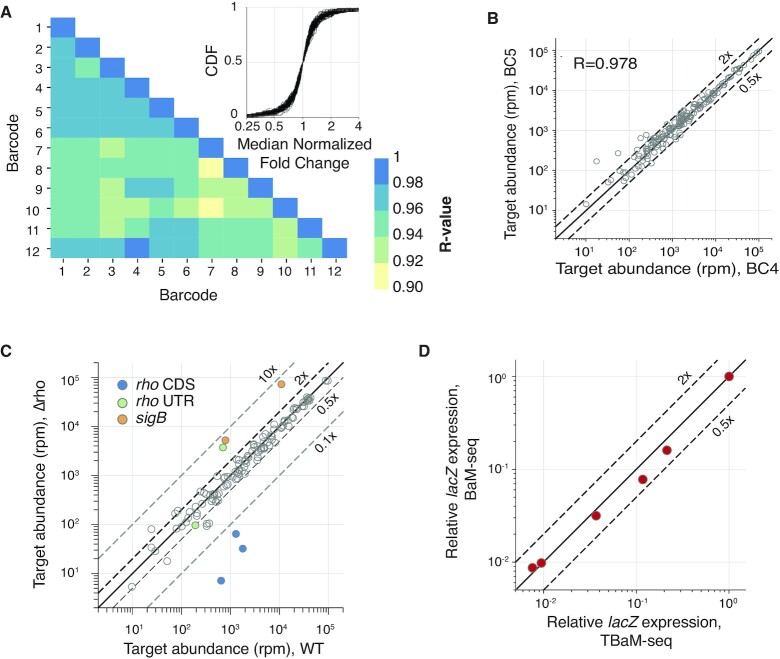
Figure 4.
Validation of TBaM-seq protocol. (A) Pearson correlation between replicates of log10-transformed reads per second-strand primer. (B) Representative example of correlation across second-strand primers between replicates. A total of 162 second-strand primers targeting 82 genes were included in this pool. The second-strand primers that show >2-fold difference in rpm between these two samples target the genes polA (0.10× between sample 4 and 5), spo0E (0.20), lysC (0.28), and glnR (0.46). polA, spo0E, and lysC are the three lowest expressed genes targeted by this second-strand primer pool (as measured by non-targeted BaM-seq). (C) Reads per million primer-mapping reads for each second-strand primer between WT and Δrho strains. Primers targeting rho CDS and UTR, as well as those targeting sigB are highlighted. (D) lacZ expression relative to sample with highest lacZ induction. For BaM-seq samples, lacZ expression is calculated as rpkm. For TBaM-seq samples, lacZ expression is calculated from median normalized reads across 12 lacZ-targeting primers.