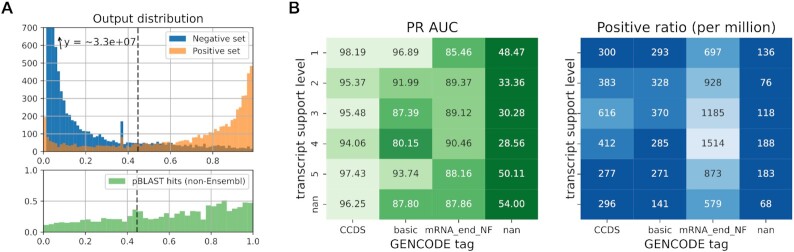
Figure 2.
Model performances on predicting translation initiation sites. (A) (top) A histogram of the model output predictions when evaluated on chromosome 2 (test set). Transcript sites are divided in a negative (blue) and positive (orange) set according to the annotations provided by Ensembl. The dotted line represents the threshold that an equal number of positive model predictions as provided by Ensembl. (bottom) The resulting coding sequences of predicted TISs were evaluated against UniProt using pBLAST. Shown are the fraction of coding sequences returning a good match in relation to the model output score. Only TISs not annotated by Ensembl (i.e., negative set) were considered. (B) Model performances were binned according to transcripts properties. PR AUC performances (left) and the ratio of positive samples in each group (right) are obtained by binning the transcripts according to transcript support level and other properties given to the annotated translation initiation site or transcript (if any, otherwise nan).