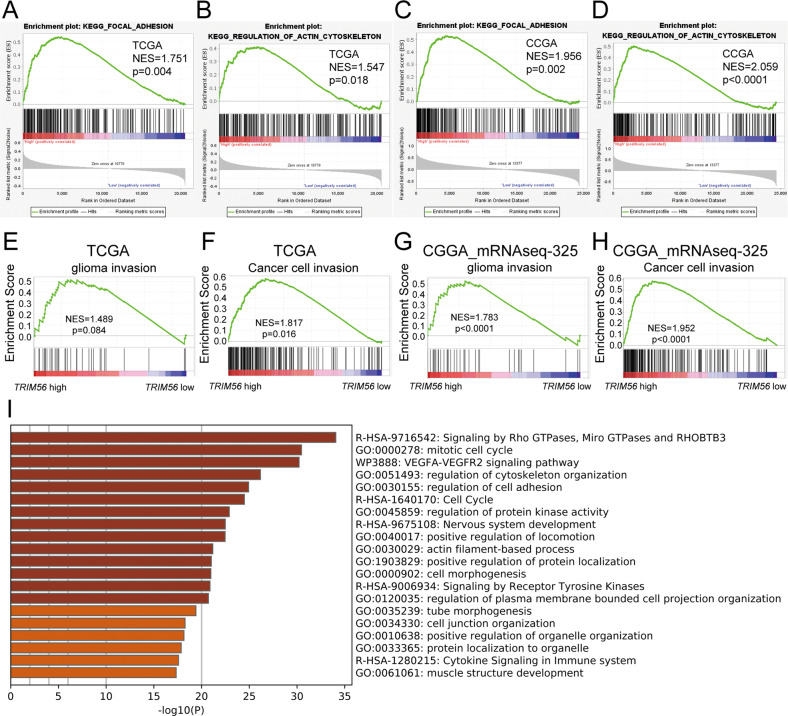
Fig. 3. Signaling pathways significantly associated with TRIM56 in glioma.
In the TCGA (A, B) and CGGA_mRNAseq_325 (C, D) cohorts, glioma samples were divided into TRIM56 high- and low-expression groups based on median TRIM56 expression. GSEA results revealed that the KEGG_FOCAL_ADHESION and KEGG_REGULATION_OF_ACTIN_CYTOSKELETON pathways were enriched in the TRIM56 high-expression group (CGGA: CGGA_mRNAseq_325 database). E–H Upon grouping the samples as described above, GSEA results revealed that glioma invasion and cancer cell invasion gene sets were enriched in the TRIM56 high-expression group. I Functional enrichment analysis of differentially expressed genes between TRIM56 knockdown cells and control cells in the Metascape platform.