FIGURE 3.
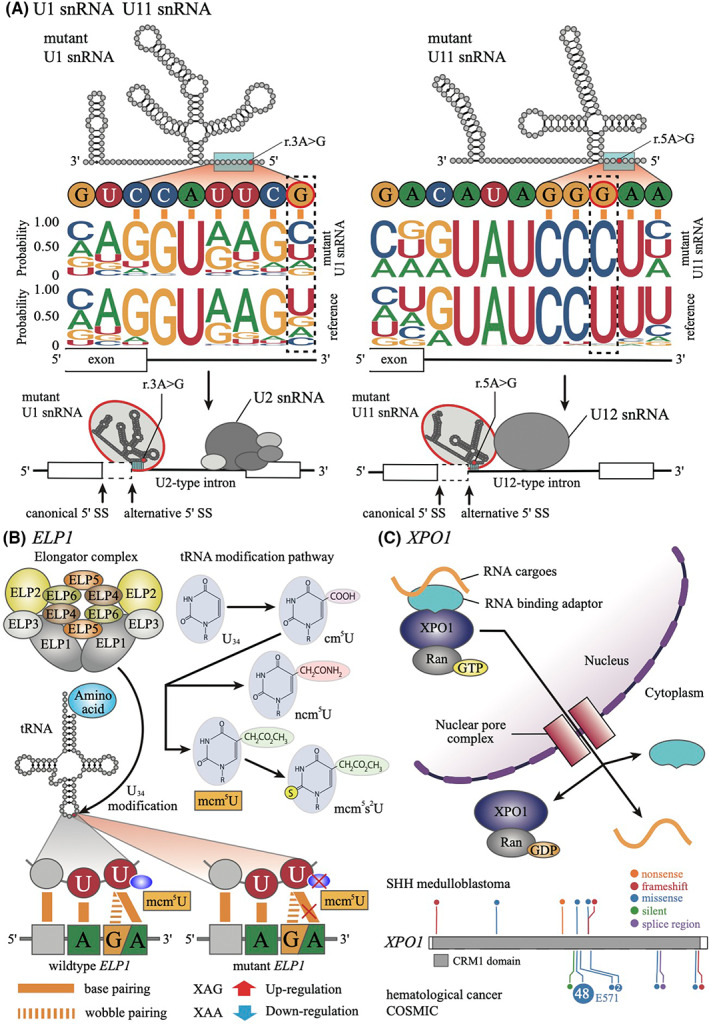
Mutations associated with RNA processing in Sonic Hedgehog (SHH) medulloblastoma. A, Schematic representation of mutations in U1 snRNA and U11 snRNA. The bases highlighted with red in the secondary structures indicate the mutational hotspots. The blue rectangle indicates the 5′ splice‐site recognition site. The upper logos are 5′ splice‐site sequences of cryptic splice sites (n = 4389) in U1 mutant SHH cases compared with U1 wildtype SHH medulloblastoma (left) and alternative splice sites (n = 5) in U11 mutant SHHδ cases compared with U11 wildtype SHHδ cases. Those splice‐site sequences were calculated using LeafCutter with RNA‐seq studies by Suzuki et al. 6 The lower logos are canonical 5′ splice‐site sequences of U2‐ and U12‐type introns from SpliceRack (http://katahdin.mssm.edu/splice/index.cgi?database=spliceNew). The lower illustrations indicate aberrant recognition of 5′ splice site by mutant U1 snRNA (left) and mutant U11 snRNA (right). SS, splice site. B, Schematic representation of tRNA modification by the elongator complex. The elongator complex modified the anticodon uridine at the wobble position (U34) indicated by the red circle and the arrow in the secondary structure of tRNA. The upper right panel shows the uridine modification in the elongator‐dependent tRNA modification pathway. ELP1 mutation causes failure of tRNA modification at the wobble position (U34), leading to an increase in AA‐ending codon usage and a decrease in AG‐ending codon usage. C, The upper panel shows the function of the nuclear export protein XPO1. The lower panel shows the mutational patterns of the XPO1 gene of SHH medulloblastoma (upper) and hematological cancer (lower) according to COSMIC v96 (https://cancer.sanger.ac.uk/cosmic). The number in the circle represents the number of cases
