Figure 5. Gene signatures distinguishing gWAT APCs and FIPs.
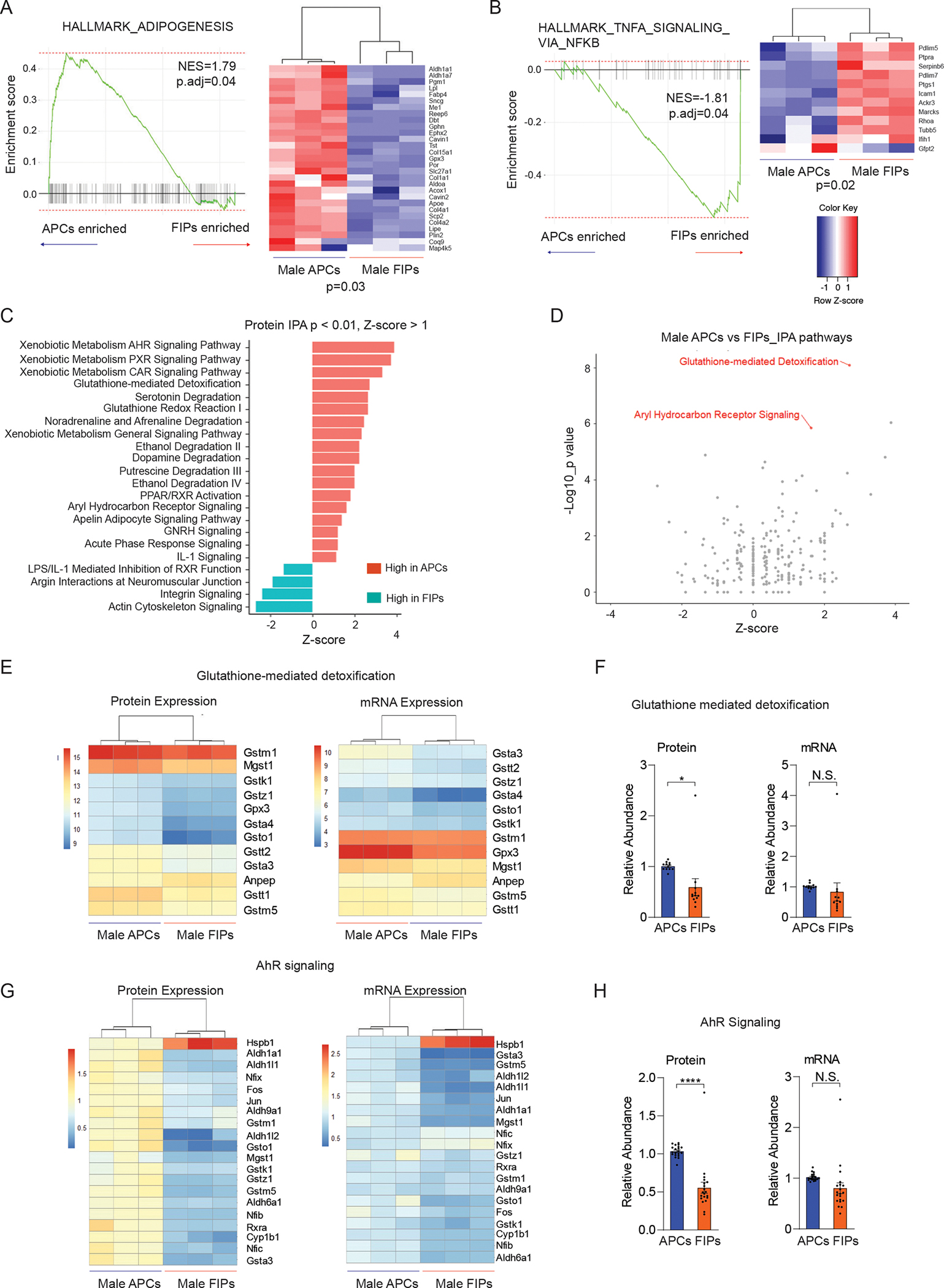
(A) Gene set enriched analysis (GSEA) based on proteomic data reveals that male APCs are enriched in an “adipogenesis” gene signature in comparison to FIPs. Heat map depicting the expression of leading-edge subset of genes.
(B) Gene set enriched analysis (GSEA) based on proteomic data reveals that male FIPs are enriched in a “TNFα signaling via NFκB” gene signature in comparison to APCs. Heat map depicting the expression of leading-edge subset of genes.
(C) Most significantly enriched pathways discriminating male APCs and FIPs identified by Ingenuity Pathway Analysis (IPA). Canonical pathways with significant coverage (p < 0.01) and a |z-score| >= 1.00 are shown.
(D) Glutathione-mediated detoxification and aryl hydrocarbon receptor (AhR) signaling were two of the most significantly upregulated pathways in male APCs when compared to FIPs.
(E) Heat map of protein levels (left) and transcript levels (right) of genes involved in glutathione-mediated detoxification in male APCs and FIPs.
(F) Expression of genes involved in glutathione-mediated detoxification were significantly enhanced in male APCs on the protein level, but not transcript level. * p < 0.05 by paired t-test. Each data point represents one gene corresponding to panel E, and y-axis shows their relative abundances.
(G) Heat map of protein levels (left) and transcript levels (right) of genes involved in AhR signaling in male APCs and FIPs. Differences in expression are more apparent on the protein level than on transcript level.
(H) Expression of genes involved in AhR signaling was significantly upregulated in male APCs on the protein level, but not transcript level. **** p < 0.00001 by paired t-test. Each data point represents one gene corresponding to panel G, and y-axis shows their relative abundances.
