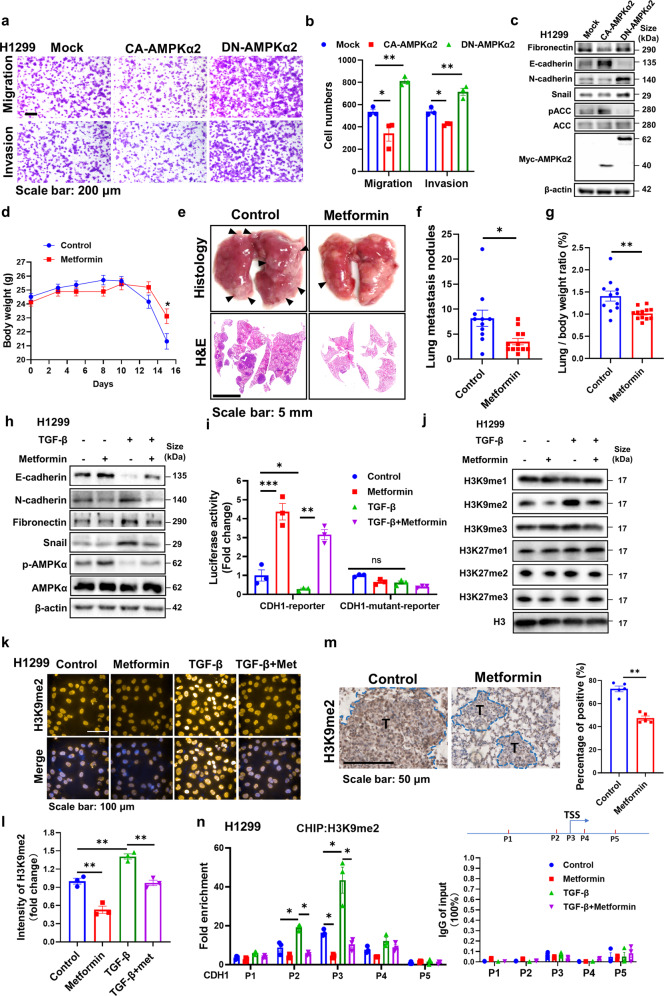
Fig. 1.
AMPK activation inhibits lung cancer metastasis and releases the repressive H3K9me2 on the promoters of epithelial genes. a, b Representative images and quantitative results of the migration and invasion in DN-AMPKα2 and CA-AMPKα2 overexpressed H1299 cells. c Western blot analysis of WCLs derived from DN-AMPKα2 and CA-AMPK α2 transfected H1299 cells. β-actin served as loading control. d Mean body weight analysis acquired at the indicated time points after orthotopic Lewis lung carcinoma (LLC) transplantation in C57BL/6 followed with metformin (250 mg/kg) treatment (n = 11, 12 for each group). e Representative images of lung nodules and H&E stained histological sections of lungs from of C57BL/6 mice acquired 15 days after orthotopic transplantation of LLC. f, g Quantitative results of lung nodules and lung/body ratio from corresponding mice. h Western blot analysis of whole-cell lysates (WCLs) derived from H1299 stimulated by TGF-β (5 ng/mL), combining with metformin treatment (1 mM) for 24 h. β-actin served as loading control. i The luciferase reporter assay containing a section of the CDH1 promoter was applied in H1299 with TGF-β (5 ng/mL) or metformin treatment (1 mM) for 24 h. CDH1 promoter luciferase values are normalized to CellTiter Glo values. j Western blot analysis of WCLs derived from H1299 stimulated by TGF-β (5 ng/mL), combining with metformin treatment (1 mM) for 72 h. H3 served as loading control. k-l Representative immunostaining images and quantitative results of the H3K9me2 intensity in TGF-β (5 ng/mL) or metformin (1 mM) treated H1299 cells for 72 h. m Representative immunohistochemical images and quantitative results of H3K9me2 in metformin treated lung cancer tissues. n CHIP-assay evaluating the recruitment of H3K9me2 to the CDH1 promoter in TGF-β (5 ng/mL) or metformin (1 mM) treated H1299 cells for 24 h with several primers around transcriptional start site (TSS). The expression levels of each IgG group of input (%) was less than 0.1%. A pattern diagram of the position of P1-P5 on the CDH1 gene promoter was shown. All error bars represent mean ± SEM. Statistical analyses were made using unpaired t-test (f, g, m) or one-way ANOVA (l) or two-way ANOVA (b, i, n) followed by multiple comparisons of fisher’s LSD tests with two tailed distribution. Statistical significance was determined at *p < 0.05, **p < 0.01, ***p < 0.001