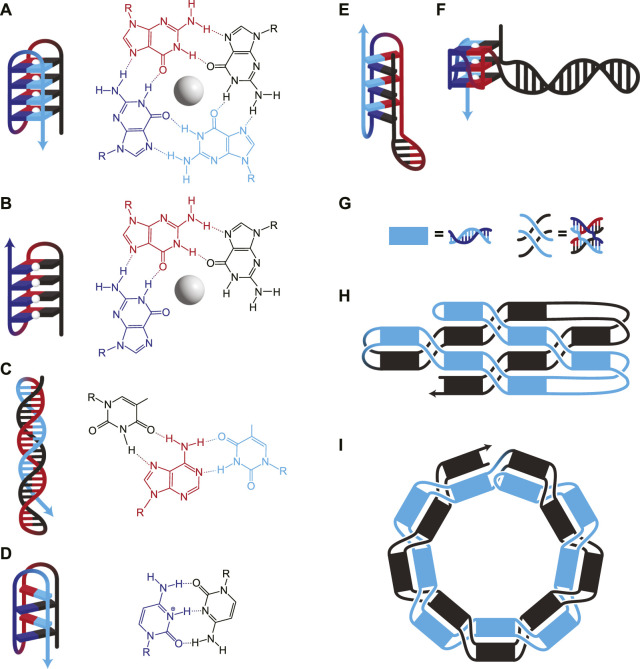
FIGURE 2.
(A–F) DNA structures involving nWC interactions. (A) G-quadruplex, where four guanines interact to form a plane around a stabilizing monovalent (typically K+) cation (white sphere). Consecutive guanines enable multiple planes which stack for additional stability. (B) G-triplex, similar to G-quadruplex, but where only three guanines interact in each plane. (C) A Hoogsteen triplex, where a WC duplex is invaded by a triplex-forming strand. The triplex-forming strand (blue) is made entirely of pyrimidines and binds only to the “central” strand (red), which is made entirely of purines. The molecular structure shows interactions on a single plane, in this case, TA*T. (D) I-motif, where intercalating pairs of C+ and C bind. Hybrids of WC and nWC structures involving (E) an i-motif with a duplex bulge and (F) a G-quadruplex with a duplex bulge where a G-quadruplex has WC base pairing within one of its loops. (G–I) Unknotted versus knotted ssDNA nanostructures. (G) Duplexes and paranemic cohesions have been visually simplified for clarity in H and I. (H) A symbolic representation of the area-filling unknotted structure. The blue section lies entirely on top of the black section, which allows the folding of this structure to occur in two thermodynamically separate steps: First, the initial formation of the helices, then the formation of the paranemic cohesions. (I) A symbolic representation of the knotted structure. At each neighbouring paranemic cohesion, the duplexes swap sides. In H and I, the figure shows a small example, but the methods have been scaled to larger structures.