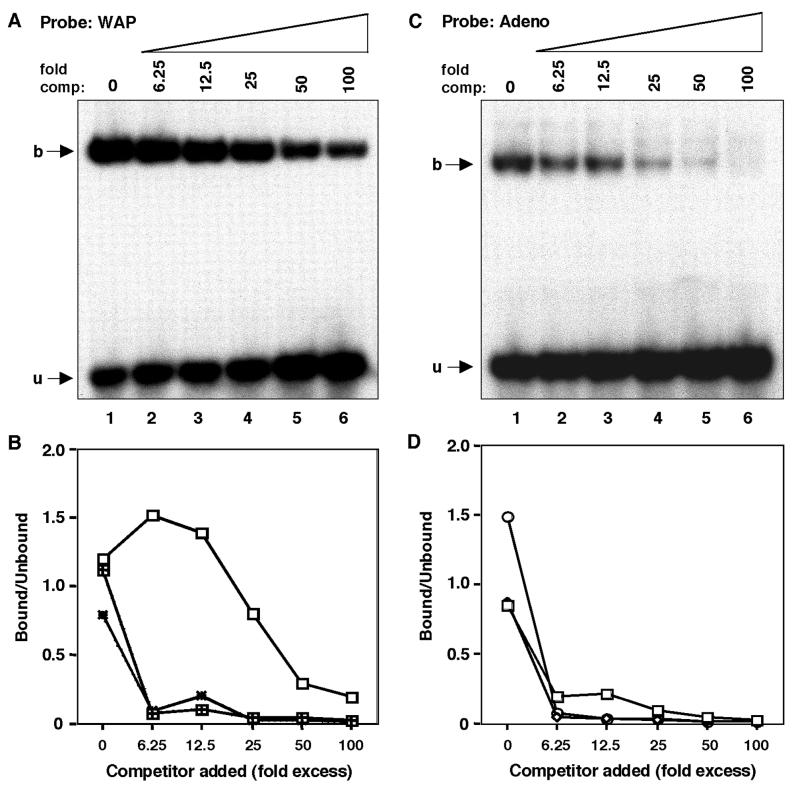
FIG. 6.
DNA-binding specificities of different NFI isoforms. (A) DNA-binding specificity of NFI-A to the palindromic motif present on the WAP distal promoter estimated by competition EMSA. The specific DNA-protein complexes (described in Materials and Methods) were made to compete with the same palindromic NFI oligonucleotide (homologous competitor) using increasing concentrations as shown (lanes 2 to 8; no competitor in lane 1). (B) Comparison of DNA-binding specificity of the three NFI isoforms to the WAP palindromic motif. The DNA-binding specificities of NFI-B and -X were also determined along with NFI-A (A) by competition EMSA, and the DNA-protein complex and free probe were quantitated by PhosphorImager. Binding specificities were determined by plotting the percentage of bound/unbound versus competitor added. Open squares represent NFI-A, squares filled with the + sign represent NFI-B, and dotted rectangles represent NFI-X. (C) DNA-binding specificity of NFI-A isoform to the palindromic motif on the adenovirus origin of replication region estimated by competition EMSA. The specific DNA-protein complexes (described in Materials and Methods) were made to compete with the same palindromic NFI oligonucleotide (homologous competitor) using increasing concentrations as shown (lanes 2 to 8; no competitor in lane 1). (D) Comparison of DNA-binding specificity of the three NFI isoforms to the adenovirus palindromic motif. The DNA-binding specificities of NFI-B and -X were also determined along with NFI-A (C) by competition EMSA, and the DNA-protein complex and free probe were quantitated by PhosphorImager. Binding specificities were determined by plotting the percentage of bound/unbound versus competitor added. Open squares represent NFI-A, open circles represent NFI-B, and the open rectangles represent NFI-X.