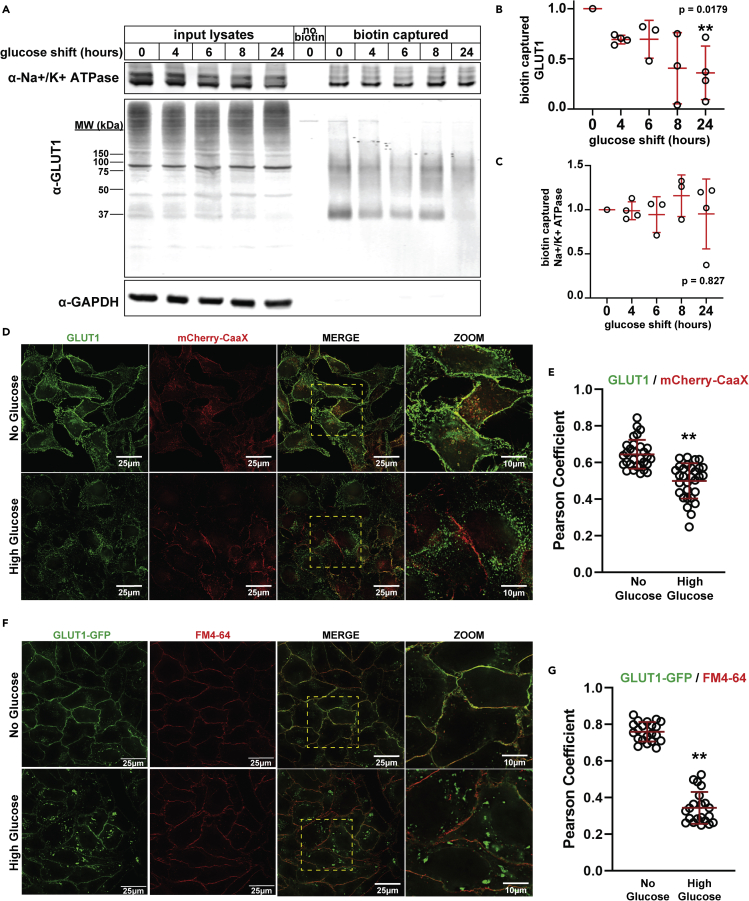
Figure 1.
Excess glucose availability promotes GLUT1 clearance from the plasma membrane
(A) HeLa cells were cultured for 24 h in media with no glucose, then switched to high glucose media (25 mM) for the indicated amount of time. Biotin-labeling was performed at the post-glucose shift time points. Following biotinylation, labeled cells were lysed and surface proteins were affinity purified with NeutrAvidin beads (Thermo Scientific). Analysis was performed by SDS-PAGE and immunoblot with antibodies that recognize GLUT1, Na+/K+ ATPase, and GAPDH.
(B,C) Quantification of captured GLUT1 (B) and Na+/K+ ATPase (C) for the experiment shown in (A) was performed over multiple biological replicates (n ≥ 3). GLUT1 measurements were taken of the whole lane using FIJI. Immunoblots for biological replicate experiments are provided in Figure S1.
(D) HeLa cells stably expressing mCherry-CaaX (red) were cultured in no glucose media for 24 h (top row) then shifted to high glucose (25 mM) for 24 h (bottom row), at which point the samples were fixed for immunofluorescence detection with GLUT1 antibody (green). Zoomed images provided in the far right column correspond to the yellow dashed-line inset boxes in the "MERGE" image to the left.
(E) Quantification of co-localization shown in (D) was measured by Pearson correlation on Softworx software (n = 30 cells), p = 3.75 × 10-8.
(F) HeLa cells stably expressing GLUT1-GFP (green) were cultured using the conditions indicated in (D). Prior to imaging, cells were pulse-labeled with FM4-64 (red), a lipophilic tracer dye that inserts into the outer leaflet of the cell membrane. Live cells were incubated on ice in 8 μM cold FM4-64 for ∼5 min before imaging. Zoomed images provided in the far right column correspond to the yellow dashed-line inset boxes in the "MERGE" image to the left.
(G) Quantification of the results shown in (F). Pearson correlation coefficient was measured using Softworx software (n = 30 cells), p = 1.69 × 10-24. For all experiments, p values were computed using a two sample Student’s t-Test in Microsoft Excel. A P value < 0.05 was considered statistically significant and is indicated by ∗∗. Data are represented as mean +/- SEM.