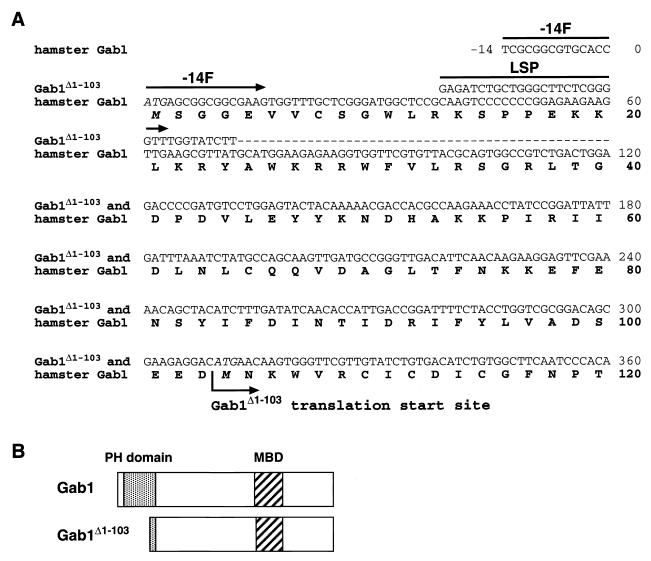
FIG. 4.
Comparison of two cDNA sequences and deduced protein sequences cloned from 10W−1L cells. (A) The nucleotide sequence of cDNA encoding Gab1 was cloned by 5′- and 3′-RACE strategy. Although hamster Gab1 is composed of 694 amino acids (2,085 nucleotide bases for the coding region), only the amino-terminal sequence of 120 amino acids (up to 360A in the nucleotide sequence) is shown here. Two cDNAs were cloned from 10W−1L, one of which (the minor product) was identical with cDNA from nontransformed SHE 83-9. A cDNA product specific to 10W−1L lacks the upstream sequence to 73G of the coding region of the original hamster Gab1 cDNA, and that sequence has been replaced by an uncharacterized sequence of at least 34 nucleotide bases, 5′-GAGATCTGCTGGGCTTCTCGGGGTTTGGTATCTT-3′ (sense), followed by the common sequence to the 3′ end. Therefore, translation of the product specific to 10W−1 starts from 310ATG (M104), instead of 1ATG (M1). A 5′ primer named −14F (5′-TCGCGGCGTGCACCATGAGCGGCGGCGAAG-3′; sense) is specific to cDNA encoding Gab1, while another 5′ primer named LSP (5′-GAGATCTGCTGGGCTTCTCGGGGTT-3′; sense) is specific to cDNA encoding Gab1Δ1-103. Dashes, nucleotide bases of Gab1Δ1-103 identical with those of hamster Gab1. (B) Comparison of the protein structures of Gab1 and Gab1Δ1-103. Gab1Δ1-103 lacks most of the amino-terminal PH domain (amino acids 14 to 116) observed in Gab1 and shares downstream structure including the MBD with Gab1.