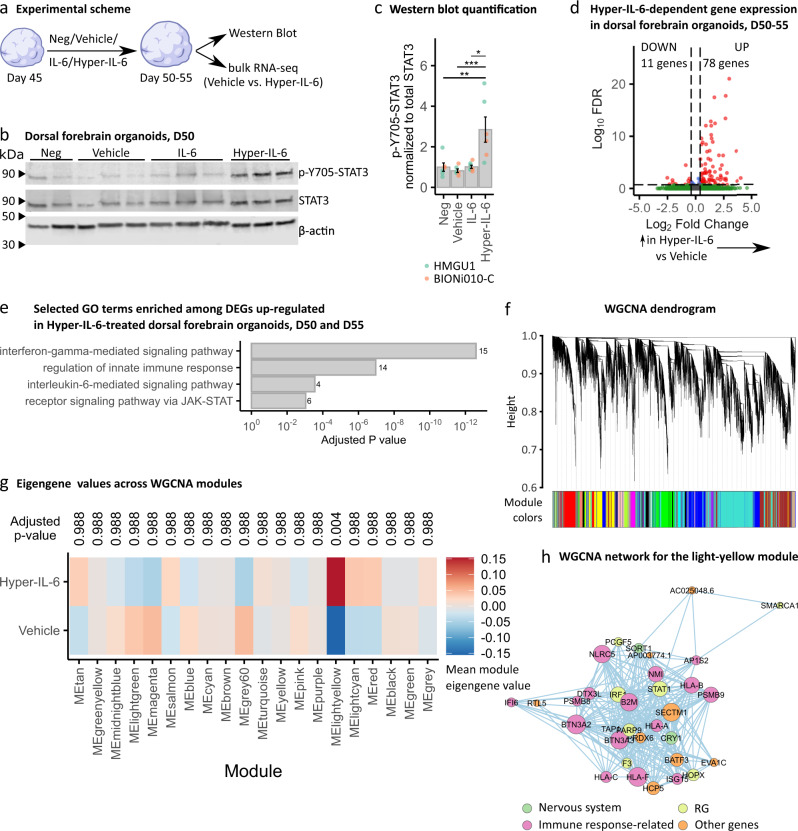
Fig. 2. Hyper-IL-6 treatment leads to the activation of JAK/STAT intracellular cascade and results in transcriptional changes in dorsal forebrain organoids.
a Scheme of the cytokine treatment in DFOs. Neg, negative control; Vehicle, vehicle control (0.1% BSA in PBS); IL-6, 8.8 ng/ml in 0.1% BSA in PBS; Hyper-IL-6, 25 ng/ml in 0.1% BSA in PBS. b Representative Western Blot of p-Y705-STAT3 (top), STAT3 (middle) and β-actin (bottom) in individual DFOs at day 50 of differentiation after 5 days of treatment. c Quantification of signal intensity of p-Y705-STAT3 relative to total STAT3 from b. Each dot represents an individual organoid sample. Color represents cell line of origin, HMGU1 (green), BIONi010-C (orange). Neg (n = 6), Vehicle (n = 6), IL-6 (n = 7), and Hyper-IL-6 (n = 6) DFOs from one batch per iPSC line. Bars represent mean, error bars represent ± SEM. Comparisons were analyzed using Aligned Rank Transform (ART) ANOVA: *p-value < 0.05, **p-value < 0.01; ***p-value < 0.001. d Volcano plot of Hyper-IL-6-dependent gene expression in DFOs at days 50 and 55 of differentiation treated for 5 to 10 days. Red dots indicate statistical significance (FDR < 0.2, absolute log2 Fold Change >0.4). Positive log2 Fold Change indicates higher gene expression in Hyper-IL-6-treated relative to Vehicle-treated DFOs. Data from n = 12 organoids per condition (two cell lines, day 50 and 55 analyzed together). e GO overrepresentation analysis (ORA) of differentially upregulated genes (FDR < 0.2, log2 Fold Change >0.4) between Hyper-IL-6- and Vehicle-treated DFOs. The x-axis displays the adjusted p-value. Numbers next to the bars represent the number of the differentially expressed genes (DEG) belonging to the GO term. Data from n = 12 organoids per condition (two cell lines, day 50 and 55 analyzed together). f Dendrogram for WGCNA obtained by clustering the dissimilarity based on consensus Topological Overlap. Each color represents a module, which contains a group of highly connected genes. A total of 20 modules were identified. g Heatmap representing mean module eigengene value by treatment condition based on WGCNA. Comparisons were analyzed using t-test, Bonferroni adjusted p-value. h WGCNA network for the light-yellow module. Color of the dots represents gene group: nervous system-enriched, green; radial glia-enriched, yellow; immune-response-related, pink; other genes, orange.