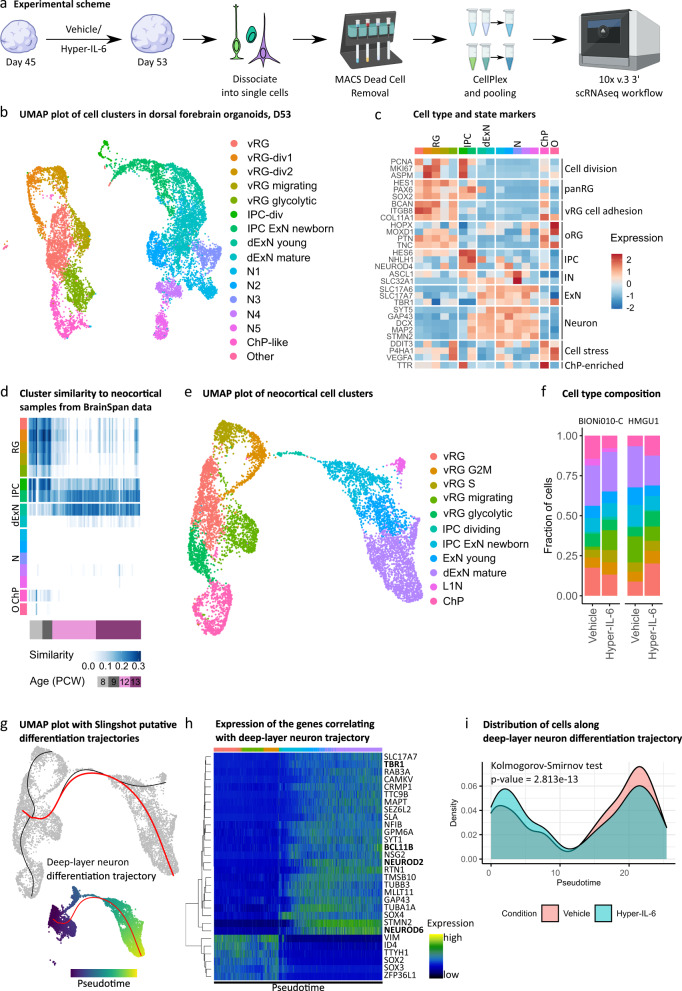
Fig. 4. Single-cell RNA sequencing of the dorsal forebrain organoids at day 53 of differentiation upon Hyper-IL-6 and Vehicle treatment.
a Schematic overview of the experimental design, including dissociation of DFOs, dead cell removal and sample multiplexing by CellPlex. b UMAP with 16 clusters with cell types labeled by color. Data from n = 2 organoids per condition. vRG, ventricular radial glia; vRG-div, dividing ventricular radial glia; IPC, intermediate progenitor cell; dExN, deep-layer excitatory neuron; N, neuron; ChP, choroid plexus. c Heatmap of gene expression level for selected known markers of neural cell types across clusters. Colors in the upper bar represent cell types from b. oRG, outer radial glia; IN, inhibitory neuron. d Heatmap of similarity metric of VoxHunt algorithm comparing organoid clusters with human neocortical RNAseq data from BrainSpan using brain regional markers obtained from Mouse Brain Atlas at E13. Colors in the left bar represent cell types from b. Data from n = 4 organoids. e UMAP with 11 clusters resulting from subclustering of cells with dorsal and medial forebrain regional identity. f Bar plots of the proportions of cell types by organoid in two cell lines. g Putative differentiation trajectories (generated by Slingshot) overlaid over UMAP, dExN differentiation trajectory in red. h, Heatmap of gene expression of genes correlating with progression through pseudotime over dExN differentiation trajectory as assessed by random forest classifier. Colors in the bar correspond to cell types from e. Hierarchical clustering of the genes shown on the left. Data from n = 4 organoids. i Density plot of cells over dExN differentiation trajectory by experimental condition. Kolmogorov-Smirnov test. Data from n = 2 organoids per condition.