Fig. 1. Cell proliferation-related and other pro-cancer signaling-related gene sets enriched to breast cancer with a high ROS score consistently in both GSE96058 and METABRIC cohorts.
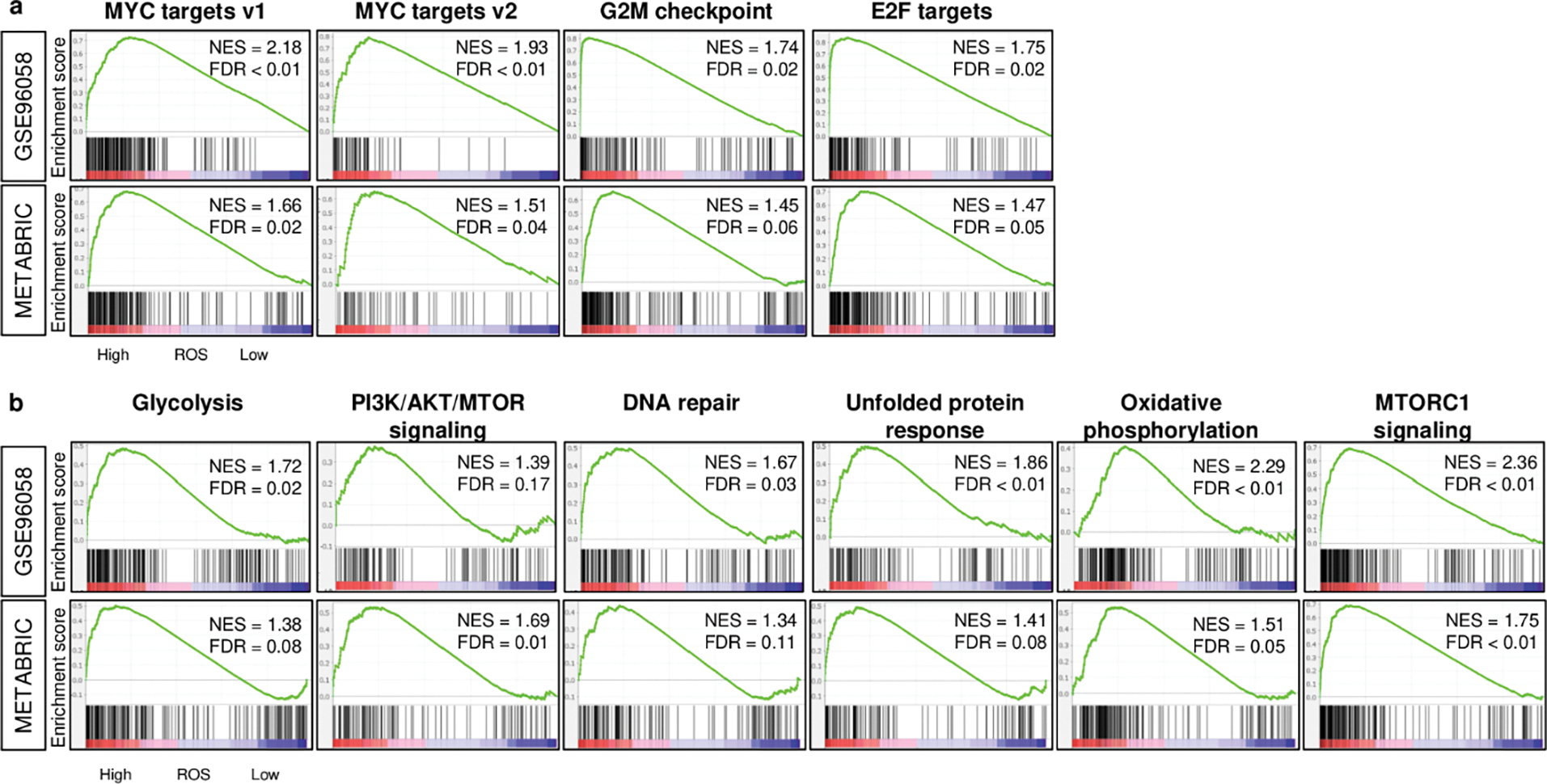
a. Enrichment plots of cell proliferation-related gene sets (MYC targets v1 and v2, G2M checkpoint, E2F targets), and b. other pro-cancerous gene sets (glycolysis, PI3K/AKT/MTOR signaling, DNA repair, Unfolded protein response, oxidative phosphorylation, and MTORC1 signaling) in the GSE96058 and METABRIC cohorts. The classical gene set enrichment analysis (GSEA) method was used to determine the NES (normalized enrichment score) and the FDR (false discovery rate) where the cut-off to divide two score groups was defined as median value
