Figure 1.
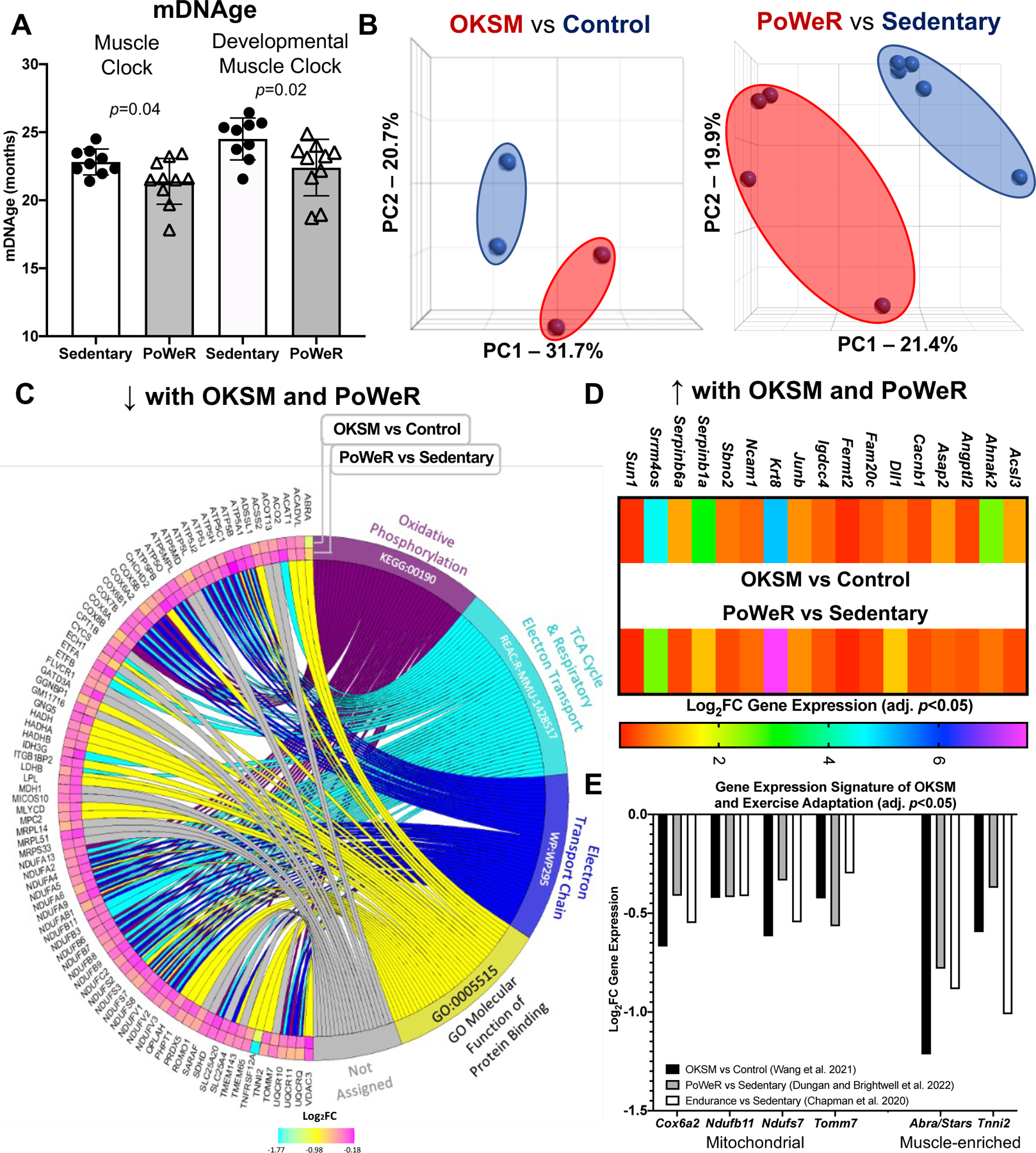
Comparison of Oct3/4, Klf4, Sox2, and Myc (OKSM) expression to late-life progressive weighted wheel running (PoWeR) in mouse muscle A DNA methylation age (mDNAge) derived from clocks developed by the Horvath laboratory applied to late-life PoWeR (n=10) and sedentary control (n=9) mouse gastrocnemius muscle B Principal component analysis (PCA) plots showing DESeq2-normalized gene count transcriptome data from OKSM expression (n=2) versus control (n=2) in mouse soleus muscle fibres (Wang et al., 2021) and the late-life PoWeR (n=4) versus the sedentary (n=5) transcriptome in soleus muscle (Dungan et al., 2022a) C Chord diagram showing significantly downregulated genes common across OKSM and late-life PoWeR in the soleus D Upregulated genes common across OKSM and late-life PoWeR in the soleus E Downregulated genes common across OKSM, late-life PoWeR, and long-term exercise training. All data reported as mean ± standard deviation
