Figure 1.
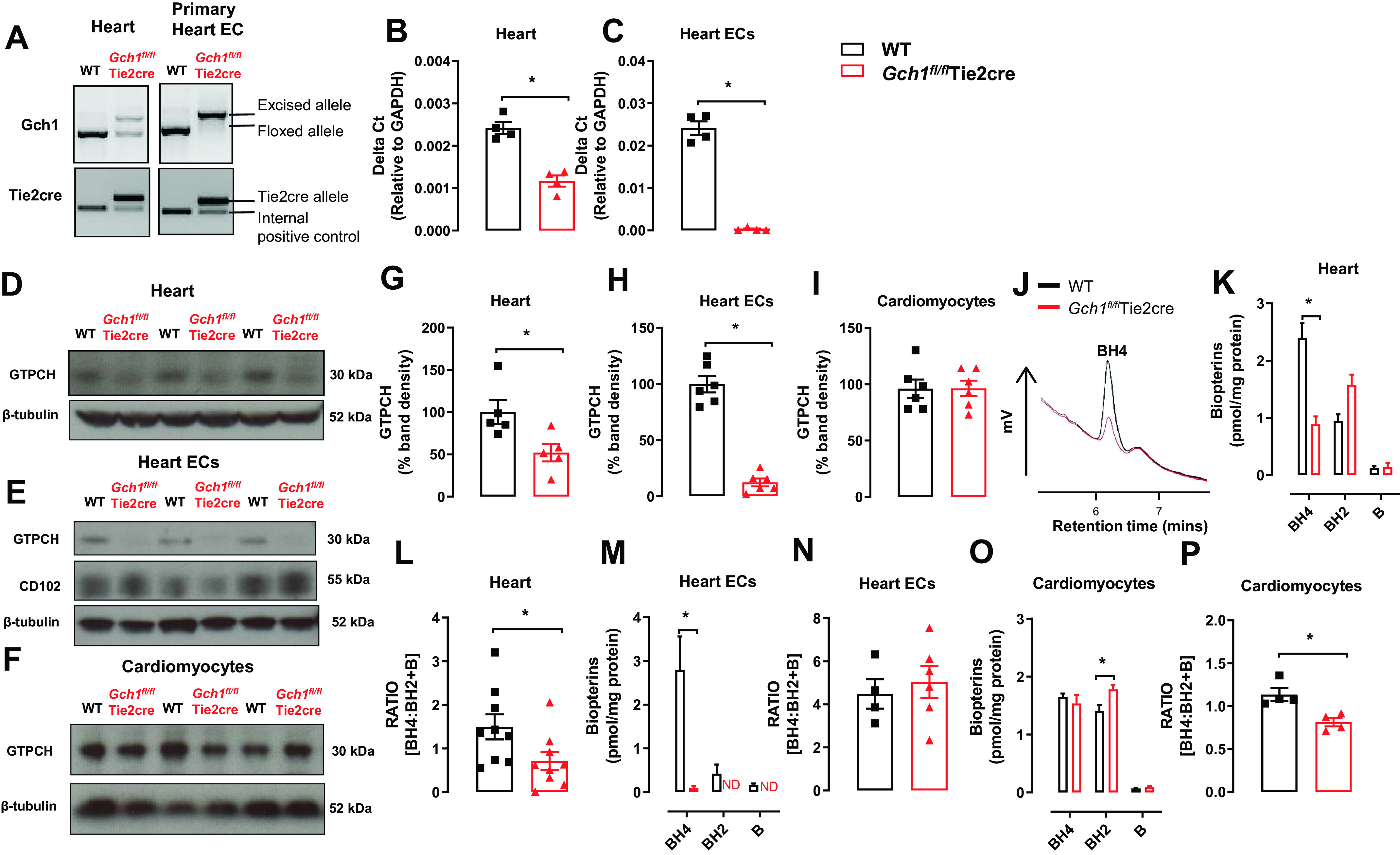
Myocardial endothelial cell targeted Gch1 deletion causes a tissue-specific decrease in Gch1 gene, GTPCH protein, and biopterin content. A: evaluation of Tie2cre-mediated excision of the loxP flanked DNA in heart tissues and primary heart endothelial cells derived from Gch1fl/flTie2cre and Gch1fl/fl [wild-type mice (WT)]. The predicted 1,030-bp product was detected in WT mice. In the presence of Tie2cre transgene a 1,392-bp knockout allele was detected, with efficient excision in primary endothelial cells from hearts. B and C: quantitative real-time PCR was used to quantify Gch1 gene expression in hearts and primary endothelial cells from hearts (*P < 0.05; n = 4 per group). D–F: representative immunoblot of GTPCH proteins in hearts, isolated primary endothelial cells, and isolated cardiomyocytes from WT and Gch1fl/flTie2cre hearts, respectively, with quantitative data, measured as percent band density in G–I: CD102 and β-tubulin were used as endothelial cell marker and loading control respectively. J: representative chromatograms of BH4 traces in hearts from WT and Gch1fl/flTie2cre mice. K and L: BH4 levels and BH4/BH2 + B ratio were reduced in hearts from Gch1fl/flTie2cre mice compared with wild-type littermates (*P < 0.05; n = 8 and 9 per group). M and N: BH4 levels were barely detectable in primary ECs from Gch1fl/flTie2cre compared with WT mice (*P < 0.05; n = 4–6 per group). O and P: BH4 levels were comparable between primary cardiomyocytes from Gch1fl/flTie2cre mice and wild-type littermates. Absolute BH2 levels in cardiomyocytes were significantly increased in Gch1fl/flTie2cre mice compared with wild-type mice, such that the BH4/BH2 and biopterin ratio was significantly reduced in cardiomyocytes in Gch1fl/flTie2cre mice (*P < 0.05; n = 4 per group). Each data point represents an individual adult male mouse.
