Abstract
Lanthanides were recently discovered as metals required in the active site of certain methanol dehydrogenases. Since then, the characterization of the lanthanome, that is, proteins involved in sensing, uptake, and utilization of lanthanides, has become an active field of research. Initial exploration of the response to lanthanides in methylotrophs has revealed that the lanthanome is not conserved and that multiple mechanisms for lanthanide utilization must exist. Here, we investigated the lanthanome in the obligate model methylotroph Methylobacillus flagellatus. We used a proteomic approach to analyze differentially regulated proteins in the presence of lanthanum. While multiple known proteins showed induction upon growth in the presence of lanthanum (Xox proteins, TonB-dependent receptor), we also identified several novel proteins not previously associated with lanthanide utilization. Among these was Mfla_0908, a periplasmic 19 kDa protein without functional annotation. The protein comprises two characteristic PepSY domains, which is why we termed the protein lanpepsy (LanP). Based on bioinformatic analysis, we speculated that LanP could be involved in lanthanide binding. Using dye competition assays, quantification of protein-bound lanthanides by inductively coupled plasma mass spectrometry, as well as isothermal titration calorimetry, we demonstrated the presence of multiple lanthanide binding sites that showed selectivity over the chemically similar calcium ion. LanP thus represents the first member of the PepSY family that binds lanthanides. Although the physiological role of LanP is still unclear, its identification is of interest for applications toward the sustainable purification and separation of rare-earth elements.
Keywords: metal, metal ion–protein interaction, proteomics, protein domain, protein purification, PepSY domain, lanthanides, methylotrophy
Abbreviations: AEX, anion-exchange; AZ3, arsenazo III; BGC, biosynthetic gene cluster; BSA, bovine serum albumin; cDNA, complementary DNA; ICP-MS, inductively coupled plasma mass spectrometry; ITC, isothermal titration calorimetry; La3+, lanthanum; LanM, lanmodulin; LanP, lanpepsy; Ln3+, lanthanide; MDH, methanol dehydrogenase; MMM, methanol minimal medium; PDB, Protein Data Bank; qPCR, quantitative PCR; XO, xylenol orange
Prior to the discovery of the first lanthanide-dependent enzyme—the methanol dehydrogenase (MDH) XoxF from Methylobacterium extorquens (1)—it was thought that lanthanides (Ln3+) do not exert a specific role in biology. In recent years, the emerging field of lanthanide biology has grown rapidly, and numerous studies have shed light on the molecular mechanisms that underlie Ln3+ uptake, trafficking, and utilization; the so-called lanthanome (2, 3, 4, 5). For methylotrophs, that is, bacteria that can grow on reduced one-carbon substrates, such as methanol, MDHs are essential for their metabolism, and the pyrroloquinoline quinone and Ln3+-dependent XoxF is particularly common (6, 7). While some methylotrophs contain only one Ln3+-dependent MDH (8), many methylotrophs, including the well-characterized M. extorquens, harbor a second type of pyrroloquinoline quinone–dependent MDH that uses Ca2+ instead of Ln3+ in its active site. This enzyme, called MxaF, is used in the absence of Ln3+. However, when lanthanides are present, the expression of mxaF is repressed, whereas xoxF expression is induced (9, 10). Given that lanthanides are largely insoluble in the environment and in phosphate-based media (11, 12), the existence of a secreted lanthanophore—a small-molecule chelator for Ln3+—has been proposed (2, 5, 10). Recently, biosynthetic gene clusters (BGCs) possibly encoding such lanthanophores have been identified in M. extorquens as well as Methylobacterium aquaticum, although the corresponding molecules have not yet been isolated and characterized (13, 14). The putative lanthanophore is likely taken up by a TonB-dependent receptor (named LutH or LanA), which has been shown to be essential for Ln3+-dependent growth (10, 15, 16). In the periplasm, the Ln3+-binding protein lanmodulin (LanM) might be involved in further Ln3+ trafficking (17, 18). Even though LanM has picomolar affinity and high selectivity (over Ca2+) for Ln3+, the protein is not essential for Ln3+-dependent growth (10), and its function is still enigmatic. In M. extorquens, an ABC transport system (including a periplasmic-binding protein) was identified that is encoded close to the TonB-dependent receptor and might be involved in the cytoplasmic import of Ln3+ (10, 16). Intriguingly, crystalline lanthanum (La3+) phosphate deposits were observed in the cytoplasm of M. extorquens by electron microscopy coupled with energy-dispersive X-ray spectroscopy (16). Extracellular (associated with outer membrane vesicles) as well as periplasmic deposits have also been described for the methylotroph Beijerinckiaceae bacterium RH AL1 (19). The exact composition, assembly, and function of these inclusions remain unclear.
Although Ln3+-dependent enzymes occur widespread, some proteins appear to be less conserved in the lanthanome, and their presence differs between the various taxa. For example, LanM as well as the putative lanthanophore BGCs are mainly present in some alphaproteobacterial methylotrophs (13, 14, 20). Furthermore, in the nonmethylotroph Pseudomonas putida, the ABC transporter is essential for growth using the Ln3+-dependent ethanol dehydrogenase (PedH); however, no homolog of the TonB-dependent receptor is present (21). These differences in the lanthanomes suggest that there are multiple mechanisms for Ln3+ sensing, uptake, and storage. Thus, additional Ln3+-interacting proteins likely exist.
Here, we investigated the lanthanome of the betaproteobacterial methylotroph Methylobacillus flagellatus KT. The bacterium employs the ribulose monophosphate cycle for one-carbon oxidation and assimilation and uses the tetrahydromethanopterin-linked pathway for formaldehyde oxidation (22, 23). M. flagellatus contains genes for both Ca2+- and Ln3+-dependent MDHs, making it possible to study the differential expression of genes in the presence and absence of Ln3+. Also, because the obligate methylotroph is unable to grow on multicarbon substrates, we expected a particularly pronounced response toward Ln3+. Furthermore, M. flagellatus does not harbor homologs of the ABC transport system, LanM, or the lanthanophore BGCs, and we thus suspected that novel proteins are involved in Ln3+ utilization.
To characterize the lanthanide response in M. flagellatus, we used a proteomics approach to assess the changes in relative protein abundance in the presence and absence of La3+. Our analysis revealed the induction of several proteins known to be involved in Ln3+ utilization, such as XoxF and the TonB-dependent receptor. Strikingly, the induced proteins also included several novel proteins. Based on bioinformatic analysis, we selected the most promising protein, Mfla_0908, for further in vitro characterization and showed that Mfla_0908 is a Ln3+-binding protein. The protein contains two PepSY domains, which so far have not been associated with Ln3+ binding. We therefore termed the protein lanpepsy (LanP), highlighting the presence of the characteristic PepSY domains. Its identification demonstrates the importance of studying diverse methylotrophs for the discovery of novel actors in the lanthanome. Our results are further of interest for applications of LanP for Ln3+ extraction, separation, and purification.
Results
The proteomic response of M. flagellatus toward La3+
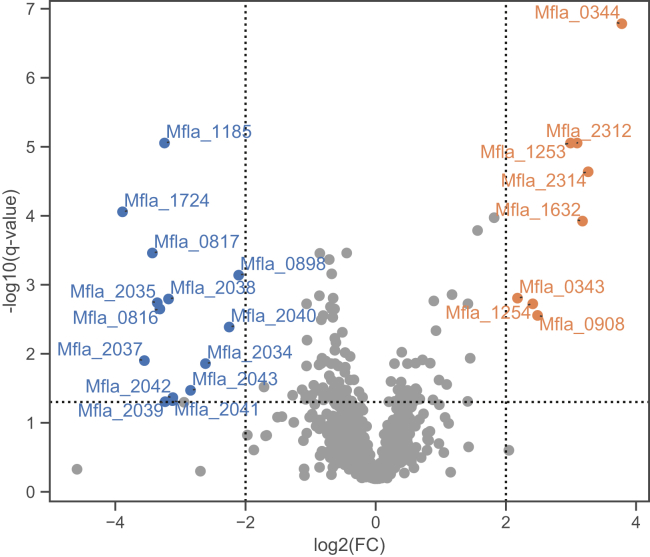
To study the physiological response of M. flagellatus toward Ln3+, we compared growth on methanol with and without LaCl3 addition using high-resolution LC–MS/MS-based label-free comparative proteomics. The experiment revealed several proteins whose relative abundance changed significantly in the presence of La3+ (Fig. 1 and Tables S1 and S2). As expected, the induced proteins included known components of the lanthanome such as the Ln3+-dependent MDH XoxF (Mfla_0344, Mfla_2314), the c-type cytochrome XoxG (Mfla_2312), and the TonB-dependent receptor involved in Ln3+ uptake (Mfla_1253). We also identified two σ54-dependent transcriptional regulators, Mfla_1254 and Mfla_0343. The gene Mfla_1254 is located directly downstream of the TonB-dependent receptor (Mfla_1253, in the same orientation) and is thus potentially involved in the regulation of its expression. Mfla_0343 is located directly upstream of xoxF (Mfla_0344, reverse orientation) and might regulate xoxF expression. Supporting this hypothesis, a putative σ54 promoter sequence was predicted upstream of xoxF by iPro54-PseKNC (24).
Figure 1.
Proteomic response of Methylobacillus flagellatus toward La3+(20 μM). Significantly changing proteins are labeled in color. The dotted lines indicate the applied fold change (|log2(FC)|>2) and q value (adjusted p value) cutoffs (q < 0.05). Only proteins detected with at least two unique peptides are shown.
Among the induced proteins were also two proteins not previously associated with Ln3+ utilization: Mfla_1632, annotated as “acriflavin resistance protein” and Mfla_0908 (=Mfla_1052; LanP) with the annotation “propeptide, PepSY and peptidase M4”. Mfla_1632 belongs to the CusA protein family, which contains inner membrane efflux pumps for heavy metal cations such as Cu+ and Ag+ (25). Strikingly, the three methionine residues (M573, M623, and M672) essential for Cu+/Ag+ binding in CusA from Escherichia coli (25) are not conserved in Mfla_1632 (replaced by His, Tyr, and Glu). Mfla_1632 is part of a predicted operon that includes an outer membrane efflux protein of the TolC family (Mfla_1630; not detected by proteomics) and a membrane fusion protein of the RND family (Mfla_1631; showing sixfold induction but only detected with one unique peptide). Together, these three proteins might form a tripartite efflux system spanning both the inner and outer membranes (26). Interestingly, the operon is also predicted to harbor a σ54 promoter sequence.
The second unknown and induced protein, LanP, is characterized by the presence of two PepSY domains and an N-terminal signal peptide predicted by SignalP (57). The PepSY domain is thought to exhibit protease inhibitory activity and is found, for example, in the propeptide of M4 metallopeptidases (27). Notably, however, the PepSY domain was recently assigned additional functions and found to be involved in uranium binding (28) and reduction of siderophore-bound iron (29).
In addition, our proteomics data revealed several proteins whose abundance decreased in the presence of La3+ (Fig. 1 and Table S2). Among the decreased proteins were expected candidates of the mxa operon such as the proteins MxaACDGIJLRS (Mfla_2034–2035, Mfla_2037–2043), which are part of the Ca2+-dependent MDH system. The MDH MxaF (Mfla_2044) also showed an eightfold decrease, although with a q value (0.0504) just slightly above the applied threshold (<0.05). Furthermore, we identified two regulatory proteins, the histidine kinase Mfla_0817 and the CitB-family response regulator Mfla_0816. Mfla_0817 shows partial homology to MxcQ from M. extorquens (only the signal transduction histidine kinase domains match), whereas the full sequence of Mfla_0816 is homologous to MxcE. In M. extorquens, the two-component systems MxcQ/MxcE and MxbD/MxbM act together to regulate mxaF and xoxF expression (9). Homologs of MxbD/MxbM also occur in M. flagellatus but do not show differential abundance in our proteomics data. Notably, in the facultative methylotroph M. extorquens, the protein levels of both two-component systems are not changing upon the addition of La3+ (10).
The proteins showing a decreased abundance upon addition of La3+ also included three proteins of unknown function: Mfla_1724, Mfla_1185, and Mfla_0898. Mfla_1724 showed the strongest decrease (15-fold) and is a small protein (21 kDa) harboring a domain of unknown function (DUF3828) and a Sec/SPI signal peptide. Mfla_1185 belongs to the thioredoxin family and might act as a protein disulfide oxidoreductase. Finally, Mfla_0898 belongs to the ferritin-like superfamily and contains a domain of unknown function (DUF455).
Growth characterization of deletion strains of induced proteins
Our proteomics data revealed several proteins induced in the presence of La3+, including a few so-far uncharacterized proteins whose role in the La3+ utilization system is unclear. To further investigate the function of these proteins, we created markerless gene deletion strains of Mfla_0343 (σ54-dependent transcriptional regulator), lanP (Mfla_0908), Mfla_1253 (TonB-dependent receptor), and Mfla_1632 (acriflavine resistance protein) and assessed the growth of each strain (as well as the WT) with and without La3+ addition (Fig. S1). We noted that the WT was growing significantly slower with La3+ than without (Δμ = 0.04 ± 0.01 h−1, p = 0.002; Δμ: difference in growth rates). A similar behavior was observed for ΔMfla_1632 (Δμ = 0.033 ± 0.004 h−1, p = 0.0001) and ΔlanP (Δμ = 0.03 ± 0.02 h−1, p = 0.03), both of which also showed a reduced growth rate when La3+ was added. However, there was no significant (i.e., with p < 0.05) difference in growth rate between the two conditions (±La3+) for the strains ΔMfla_0343 and ΔMfla_1253. This lack of a growth defect in the presence of La3+ suggests that these strains might no longer be able to sense or utilize La3+. For Mfla_1253—the TonB-dependent receptor—it has already been shown in M. extorquens that the ortholog is essential for Ln3+ uptake (10, 16). In its absence, Ln3+ cannot be sensed, and xoxF is thus not expressed. Our data therefore are consistent with the importance of the TonB-dependent receptor for Ln3+ utilization also in M. flagellatus. For Mfla_0343—the σ54-dependent transcriptional regulator encoded next to xoxF—the absence of a growth defect in the presence of La3+ further supports the hypothesis that the protein is involved in the regulation of xoxF expression.
Although ΔlanP showed no growth phenotype compared with the WT, we tested if the strain was still able to modulate mxaF/xoxF expression in response to La3+ using RT–quantitative PCR (qPCR) (Fig. S2). Indeed, in the presence of La3+, xoxF (Mfla_0344) was fully expressed, whereas mxaA (Mfla_2038), which was used as a proxy for the expression of the mxa cluster, was repressed. In the tested condition, the function of LanP hence seems to not be directly linked to Ln3+ sensing or uptake.
The PepSY protein LanP is a putative metal-binding protein linked to methylotrophy
Although the deletion of lanP (Mfla_0908) did not result in a growth phenotype, several properties of this protein triggered our interest. Its small size (19 kDa), predicted periplasmic localization (based on the presence of a Sec/SPI signal peptide, Fig. 2A), as well as the presence of a large number of negatively charged residues led us to speculate that the protein could be involved in Ln3+ binding. In addition, the presence of two PepSY domains (Pfam PF03413, Fig. 2A) in a small protein was reminiscent of UipA, a protein that has recently been identified as a high-affinity uranium- (UO22+) and iron- (Fe3+) binding protein in several Microbacterium strains (28). Notably, UipA (Protein Data Bank [PDB] ID: 7ATH) was also the second-best hit in an HHpred search (30) of the LanP sequence against the PDB (the best hit being PDB ID 4EXR, a putative lipoprotein). In UipA, the highly negatively charged PepSY domains form a domain-swapped dimer and are involved in the binding of multiple metal ions. However, in contrast to UipA, which is membrane anchored with a single-pass transmembrane helix, LanP is predicted to be periplasmic and does not encode predicted transmembrane sequences.
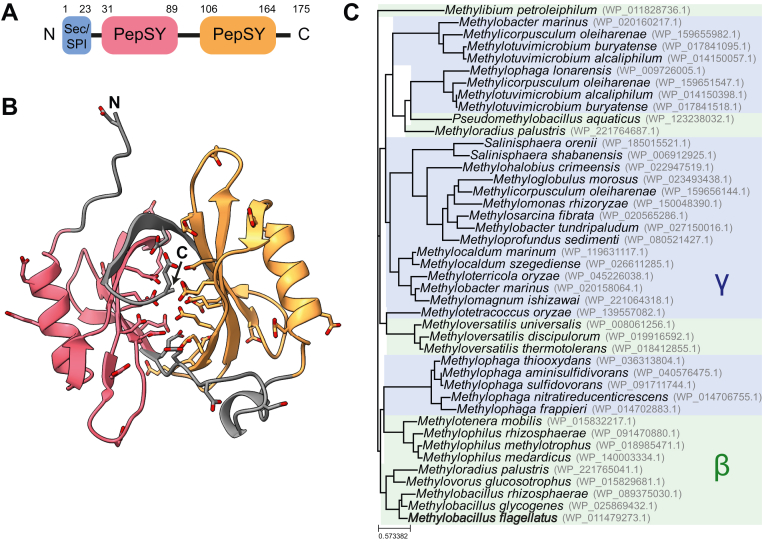
Figure 2.
Bioinformatic analysis of LanP (Mfla_0908).A, protein domain arrangement in LanP. Sec/SPI depicts the N-terminal signal peptide as predicted by SignalP 6.0 (57). The PepSY domains were annotated based on an NCBI conserved domain search (58). B, predicted 3D structure of the LanP monomer generated using AlphaFold (31). The (unfolded) signal peptide (residues 1–23) has been removed from the model. The colors indicate the two PepSY domains as depicted in A. Negatively charged residues (Glu and Asp) are shown as sticks. C, phylogenetic tree of LanP homologs. Sequences were selected by a NCBI BLAST search against the RefSeq Select database. Relatively strict criteria were applied to filter the sequences to only obtain homologs with potentially the same function (≥85% query coverage, E value ≤10−20). Betaproteobacteria are shaded in green, whereas gammaproteobacteria are shaded in blue.
After analyzing the primary sequence of LanP, we next generated a de novo 3D structural model using AlphaFold (Deepmind) (31) (Fig. 2B). The predicted structure had a high accuracy score (pLDDT values above 90, except for termini and the unfolded signal peptide). In the predicted structure, both PepSY domains adopt the same fold, and they arrange into a pseudodimeric structure. The structure of the individual domains was similar to the PepSY domains in the crystal structure of UipA (28), although the relative positioning of the 2 domains is different (Fig. S3). In the case of LanP, the β-sheets present in each PepSY domain form a β-barrel-like structure instead of the concave β-sheet surface present in UipA. Strikingly, the lumen of the barrel of LanP is lined with 19 negatively charged glutamate and aspartate residues (shown as sticks in Fig. 2B), resulting in a low protein pI of 4.6. We speculated that these residues might be involved in metal binding, similarly as described for UipA and LanM (18, 28). In the case of LanM, Ln3+ coordination requires four to five carboxylates and one backbone carbonyl per ion; therefore, the lumen of LanP might even harbor several metal-binding sites.
To further investigate the hypothesis that LanP functions as a metal-binding protein, we determined the phylogenetic distribution of LanP. After performing a BLAST search in the National Center for Biotechnology Information (NCBI) RefSeq Select database, we applied relatively strict criteria to further filter the sequences for homologs with potentially the same function (≥85% query coverage, E value ≤10−20). As indicated by the "Methylo" prefix of most of the species names in the resulting phylogenetic tree (Fig. 2C), it was immediately evident that most strains are probably methylotrophs. Indeed, checking for the presence of XoxF/MxaF homologs (the two proteins cannot be distinguished solely by BLAST) confirmed that all strains are predicted to encode at least one MDH. Furthermore, most strains seem to use the ribulose monophosphate cycle for assimilation of methanol, as genes for 3-hexulose-6-phosphate synthase (Hps) and 6-phosphate-3-hexuloisomerase (Phi) could be identified in all strains except for Methylibium petroleiphilum and the three Methyloversatilis species. Interestingly, none of the strains shown in the phylogenetic tree (Fig. 2C) encoded a homolog of LanM. In addition, all strains in the tree belong to the beta- or gammaproteobacteria. However, when applying less rigorous cutoffs and using DELTA-BLAST (32), more distant homologs (mainly annotated as “PepSY domain–containing protein”) could also be detected in M. extorquens AM1 (MexAM1_META1p4353) and in numerous non-methylotrophs belonging primarily to the phyla Firmicutes or Fusobacteriota. Notably, even these distant homologs contain a high number of negatively charged amino acids, indicating that they might be functionally relevant.
The increased abundance of LanP in the presence of La3+, the similarity to the uranium-binding protein UipA, as well as the exclusive presence of close LanP homologs in methylotrophs provided evidence that the protein might bind Ln3+ and thus play a specific role in Ln3+-dependent methylotrophy. Of note, the Ln3+-binding protein LanM also binds several actinides (33, 34, 35), thus making the connection between LanP and the uranium (an actinide) binding by UipA even more interesting. All these findings encouraged us to further experimentally characterize LanP.
In vitro characterization of LanP reveals multiple Ln3+-binding sites
To characterize LanP (Mfla_0908) in vitro, we heterologously expressed and purified the protein from E. coli. For initial expression, we removed the signal peptide (residues 1–23) and fused the sequence with an N-terminal His10 tag. Affinity purification followed by size-exclusion chromatography (in the absence of La3+) unexpectedly revealed the formation of two main species of largely different molecular weights (Fig. S4). The main peak seemed to originate from protein oligomers with estimated molecular weights of ∼550 kDa, thus corresponding to approximately 30 monomers of LanP (19 kDa each). To ensure that the high–molecular-weight structure was not an artifact, we expressed LanP without affinity tag and purified the protein using several chromatographic steps (Fig. S5). Strikingly, the large protein complexes were not observed anymore during size-exclusion chromatography and LanP eluted as a dimer (Fig. S5A). We therefore used untagged LanP for all further in vitro characterizations.
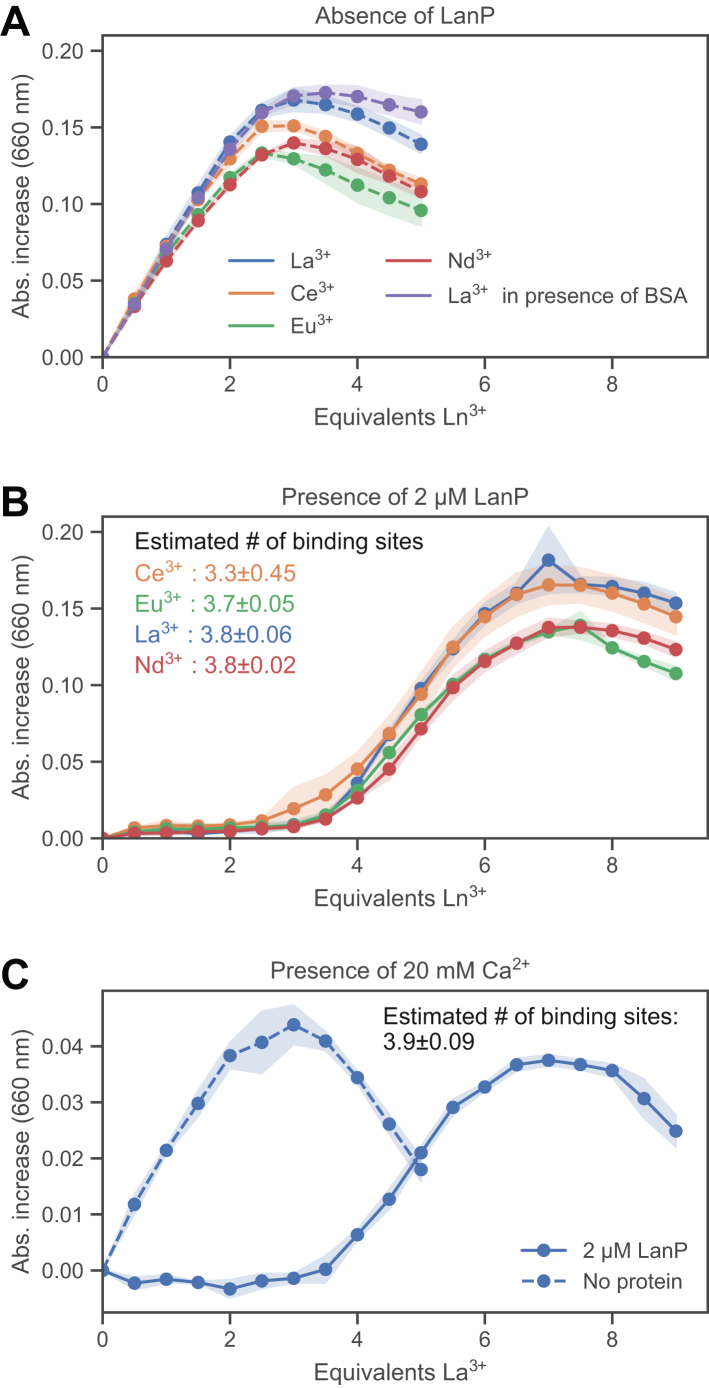
To test our hypothesis that LanP is a Ln3+-binding protein, we performed an in vitro competition assay using the metal-chelating dye arsenazo III (AZ3). Multiple assays based on this dye have previously been used to quantify Ln3+ (36, 37, 38), more recently also from culture supernatants (39). In our assay, purified LanP competed with AZ3 for the binding of Ln3+. The absorption at 660 nm was used to monitor the binding of Ln3+ to the dye during the titration. In the absence of LanP as well as in the presence of the control protein bovine serum albumin (BSA), a titration with the four lanthanides La3+, Ce3+, Eu3+, and Nd3+ revealed a linear increase of the absorbance until the dye was fully saturated (Fig. 3A). However, when 2 μM LanP was present, the increase in absorbance was delayed until 3 to 4 molar equivalents of Ln3+ were added (Fig. 3B). These results suggest that LanP has up to four Ln3+-binding sites with affinities that are higher than the one of AZ3. Dissociation constants for complexes between Ln3+ and AZ3 have been determined in the low micromolar to low nanomolar range (40, 41, 42).
Figure 3.
Dye competition assay using purified LanP (Mfla_0908).A, control assays performed in the absence of LanP or in the presence of bovine serum albumin (BSA) (2 μM) using different lanthanides for titration. The absorbance increase at 660 nm indicates the binding of Ln3+ to arsenazo III (10 μM). The equivalents Ln3+ shown in the x-axis are based on the 2 μM LanP used in B. The shaded areas represent the standard deviation of three independent assays. B, competition assay between 2 μM LanP and 10 μM arsenazo III. The delay in absorbance increase compared with the control (A) is due to the binding of Ln3+ by LanP. The number of binding sites was estimated based on the molar equivalent Ln3+ required to reach 15% of the maximum absorbance, indicating saturation of the binding sites on LanP. The values are reported as mean ± standard deviation (n = 3). C, competition assay similar as in B, but La3+ was titrated into a solution containing in addition 20 mM CaCl2.
To determine if LanP can distinguish between La3+ and the chemically similar Ca2+ ion (12), we performed the competition assay in the presence of a 2500-fold excess of CaCl2 (compared with the La3+ concentration at saturation of LanP). Strikingly, the presence of Ca2+ did not influence the binding of La3+ to LanP and still ∼4 equivalents were required to fully saturate the protein (Fig. 3C).
We further tested the ability of LanP to bind the nonlanthanide metal ions Fe2+, Fe3+, and Zn2+ in competition assays. Notably, zinc and iron are the two most abundant transition metals in bacterial (E. coli) cells (43). Since the range of metal ions that can be used in such assays is limited by the binding of the ions to the indicator dye in our buffer system, we replaced AZ3 with xylenol orange (XO) in the case of Zn2+. The titrations with these nonlanthanide metal ions did not show a shift in the linear increase of the absorbance (Fig. S6), indicating that these ions did not bind to the protein or with affinities much lower than for the indicator dyes. The binding sites in LanP thus seem to be selective for Ln3+.
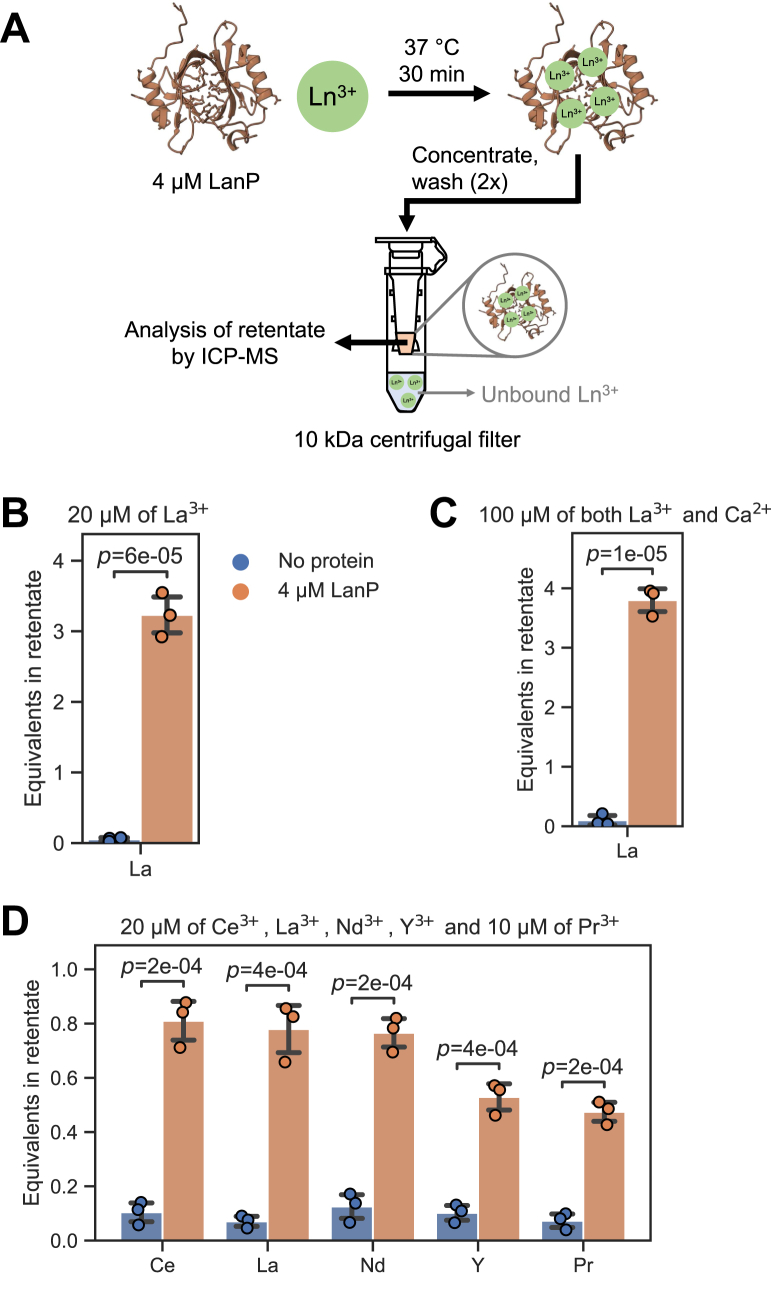
To validate the findings from the competition assay, we performed experiments to directly measure the protein-bound Ln3+ using inductively coupled plasma mass spectrometry (ICP-MS). After incubation of a Ln3+ solution with LanP, we used centrifugal filters (10 kDa molecular-weight cutoff) to separate the protein from unbound metals (Fig. 4A). After two washing steps, lanthanides in the retentate were quantified by ICP-MS. When 4 μM LanP was incubated with 20 μM of La3+, 3.2 ± 0.3 equivalents of La3+ could be recovered from the protein fraction (Fig. 4B). This value is only slightly lower than the ∼4 binding sites determined in the aforementioned competition assay. A negative control without protein showed almost no retention of La3+ in the filter (0.05 ± 0.03 equivalents; Fig. 4B). Next, we incubated the protein with a mixture of 100 μM La3+ and 100 μM Ca2+ to confirm the selectivity of the protein. Similar as in the competition assay, the protein was able to bind 3.8 ± 0.2 equivalents of La3+, even in the presence of Ca2+ (Fig. 4C). In addition, we used a mixture of the five rare-earth elements Ce3+, La3+, Nd3+, Y3+, and Pr3+ (all 20 μM, except Pr3+, which was 10 μM) to determine if certain elements show preferential binding to LanP. Equal binding was observed for Ce3+ (0.81 ± 0.09 equivalents), La3+ (0.8 ± 0.1 equivalents), and Nd3+ (0.77 ± 0.06 equivalents) (Fig. 4D). Protein-bound Y3+ (0.53 ± 0.06 equivalents) and Pr3+ (0.47 ± 0.04 equivalents) were slightly less abundant in the retentate and might thus be bound with lower affinities.
Figure 4.
ICP–MS analysis of retentates obtained by ultrafiltration of LanP (Mfla_0908) incubated with different rare-earth elements.A, schematic illustration of the binding assay and the removal of unbound metals by centrifugal ultrafiltration. B–D, quantification of rare-earth elements in retentates by ICP–MS. In the negative control, no protein was present (blue bars). For the assays with protein (orange bars), 4 μM LanP was incubated with (B) 20 μM LaCl3, (C) 100 μM LaCl3 and 100 μM CaCl2, and (D) 20 μM of each CeCl3, LaCl3, NdCl3, YCl3, and 10 μM PrCl3. Each assay was repeated three times, and the error bars depict the standard deviation. ICP–MS, inductively coupled plasma mass spectrometry.
To quantify the binding affinities of LanP for different metals, we used isothermal titration calorimetry (ITC) and titrated the lanthanides La3+, Nd3+, and Eu3+ as well as the nonlanthanides Ca2+ and Fe3+ (all at 5 mM) into a solution of 50 μM LanP (Table S3). For the lanthanides, the binding isotherms showed a characteristic biphasic shape, and they could be best fitted using a model with two sets of (independent) sites. The model suggested that for all three lanthanides, the first set of sites had a similar stoichiometry of 6 (NLa = 6.1 ± 0.2, NNd = 6.1 ± 0.3, and NEu = 6.4 ± 0.3) with an affinity of 1 μM (Kd,La = 1.1 ± 0.1 μM, Kd,Nd = 1.1 ± 0.3 μM, and Kd,Eu = 1.1 ± 0.2 μM). This set of sites likely corresponds to the binding sites observed in the competition assay, although it is curious why two additional binding sites are obtained compared with the ∼4 sites determined by the competition assay or ICP-MS. The second set of sites showed a stoichiometry of 0.2 (NLa = 0.2 ± 0.2, NNd = 0.2 ± 0.4, and NEu = 0.2 ± 0.3) with a dissociation constant of 5 μM (Kd,La = 5 ± 1 μM, Kd,Nd = 5 ± 1 μM, and Kd,Eu = 5 ± 0.7 μM). This set of sites might be interpreted as an additional binding site that is only available in a fraction (20%) of the LanP molecules. Alternatively, there could be additional processes that contribute to the isotherms, such as metal-induced conformational changes or (de)multimerization of LanP.
The titration of Ca2+ resulted only in a weak signal, leading to a large uncertainty in the affinity (Kd = 14 ± 11 μM with N = 4.3 ± 0.4). For Fe3+, the isotherm showed only noise (Table S3), and no fitting was possible, demonstrating that LanP does not bind Fe3+. While a more detailed analysis will be necessary to fully elucidate the dynamics of Ln3+ binding by LanP, the ITC measurements confirm the binding of up to six lanthanides and support the selectivity of Ln3+ over Ca2+ and Fe3+.
Discussion
The proteomic analysis of M. flagellatus comparing growth with and without La3+ revealed both known as well as novel proteins involved in the lanthanome. The most induced proteins included the acriflavine resistance protein Mfla_1632 (ninefold induced) that is part of a tripartite efflux system spanning both the inner membrane and the outer membrane. Similar export systems are known to be involved in the secretion of a wide range of (toxic) compounds, such as antibiotics, heavy metals, and proteins (26, 44). It thus seems plausible that the identified transport system might be involved in the export of excess Ln3+, for example, for detoxification purposes, or in the export of a lanthanophore for subsequent uptake of chelated Ln3+ via the TonB-dependent receptor. Notably, a homolog of this protein is also present in M. extorquens, although it is not induced in the presence of La3+ (10).
The regulatory networks involved in the lanthanide switch leading to the differential expression of the XoxF/MxaF MDHs are complex and still not fully understood (45). Here, we identified an additional σ54-dependent regulatory system that includes the two proteins Mfla_1254 and Mfla_0343, both showing fivefold induction in the presence of La3+. This system likely acts together with the two two-component systems MxcQ/MxcE and MxbD/MxbM, previously described in M. extorquens AM1 (9), which are also present in M. flagellatus. So far, σ54 regulators have not been described in the context of the lanthanome, and their discovery further highlights the complexity and diversity of the regulatory mechanisms used by different methylotrophs. Notably, M. extorquens also encodes σ54-dependent regulators; however, potential homologs of Mfla_1254 or Mfla_0343 do not appear to be associated with known proteins of the lanthanome.
In vitro characterization of LanP, a PepSY domain–containing protein showing sixfold induction in the presence of La3+, demonstrated the existence of multiple selective binding sites for Ln3+ with affinities of 1 μM. Although the PepSY domain has recently been associated with uranium and iron binding (28), Ln3+ binding has so far not been described for this protein family. LanP thus represents—after LanM (17, 18)—only the second characterized nonenzymatic protein involved in Ln3+ binding in methylotrophy. Although LanM and LanP do not show similarity in terms of their primary sequence or (predicted) structure, they share several functional properties: both are small and periplasmic proteins that harbor multiple Ln3+-binding sites that show selectivity over Ca2+. Both proteins are not essential for Ln3+-dependent growth on methanol, and no phenotype of the deletion strains could be identified so far (10). As M. flagellatus does not encode a homolog of LanM, its function might be replaced by LanP. Both proteins might thus be involved in similar processes that are not growth limiting under the conditions tested so far.
Ln3+-binding proteins are attractive targets for applications in extraction, separation, and recycling of Ln3+ (46). The fact that proteins are nontoxic, biodegradable, and that they can relatively easily be engineered or evolved for improved properties (e.g., affinity, selectivity, stability) provides an advantage compared with other (synthetic) Ln3+ chelators. In case of LanM, its ability to selectively extract rare-earth elements from industrial feedstocks has already been demonstrated (47). LanM further proved to be surprisingly stable at low pH and high temperatures, which are important features for industrial applications (47). Here, we applied ultrafiltration using protein concentrators to validate Ln3+ binding to LanP. The fact that protein-bound Ln3+ could be retained even after several washing steps further served as a proof of concept for the application of LanP for Ln3+ purification.
In summary, the work presented here has led to the identification of several new players in the lanthanome of M. flagellatus: the Ln3+-binding protein LanP, a tripartite efflux system (acriflavine resistance protein), as well as two σ54-dependent transcriptional regulators. Future characterization of LanP will be of interest to determine the mechanistic and structural basis for binding and selectivity, which could also benefit (bio-)technological applications.
Experimental procedures
Cultivation of M. flagellatus
M. flagellatus strains were cultured in 20 ml minimal medium in 100 ml baffled flasks at 37 °C. The methanol minimal medium (MMM) contained 125 mM methanol as carbon source, mineral salts (1.62 g/l NH4Cl, 0.2 g/l MgSO4·7H2O), phosphate buffer (2.4 g/l K2HPO4, 1.08 g/l NaH2PO4·2H2O), iron solution (15 mg/l Na2EDTA·2H2O, 3 mg/l FeSO4·7H2O), and trace elements (4.5 mg/l ZnSO4·7H2O, 3 mg/l CoCl2·6H2O, 0.64 mg/l MnCl2, 1 mg/l H3BO3, 0.4 mg/l Na2MoO4·2H2O, 0.3 mg/l CuSO4·5H2O, 3 mg/l CaCl2·2H2O). For conditions containing La3+, LaCl3 was added at a final concentration of 20 μM.
Proteomic analysis of M. flagellatus
For a detailed step-by-step protocol, refer to Ref. (48). Briefly, M. flagellatus KT was grown as independent precultures and main cultures with or without 20 μM LaCl3. From the main cultures, cells (corresponding to 4 ml with absorbance at 600 nm of 1) were harvested by centrifugation followed by washing with 10 mM MgCl2. The obtained cell pellets were shock-frozen in liquid nitrogen and stored at −80 °C until further processing. In total, five independent biological replicates were generated for each condition. Bacterial cell pellets were dissolved and lysed as described previously (10, 48, 49) in lysis buffer containing 100 mM ammonium bicarbonate, 8 M urea, and 1× cOmplete EDTA-free protease inhibitor cocktail (Roche). Lysis was achieved by indirect sonication (3× 1 min, 100% amplitude, 0.8 cycle time) in a VialTweeter (Hielscher). Insoluble parts were removed by centrifugation (13,000g, 15 min, 4 °C), and supernatant protein concentration was determined using the Pierce BSA assay kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Disulfide bonds were reduced by the addition of 5 mM Tris(2-carboxylethyl)phosphine and incubation for 30 min at 37 °C. Cysteine residues were alkylated by adding 10 mM iodoacetamide and incubation for 30 min in the dark at room temperature. Afterward, samples were diluted 1:5 with fresh 50 mM ammonium bicarbonate to reduce the urea concentration below 2 M. Proteins were digested overnight at 37 °C and shaking at 300 rpm using sequencing grade modified trypsin (Promega) at an enzyme to protein ratio of 1:50. After digestion, the trypsin was heat inactivated (95 °C, 5 min), and formic acid was added to an approximate concentration of 1%. Peptide samples were centrifuged (20,000g, 10 min), and the supernatant was taken and desalted using Sep-Pak Vac C18 reversed phase columns (Waters) as described previously (48, 49) and dried under vacuum. Prior to LC–MS/MS analysis, the samples were resolubilized in 3% acetonitrile containing 0.1% formic acid.
MS analysis of peptide samples was performed on an M-Class ultraperformance liquid chromatography system (Waters) coupled to an Orbitrap Fusion (Thermo Fisher Scientific) equipped with a digital PicoView nanospray ion source (New Objective). Chromatographic separation of peptides was performed using 0.1% formic acid for solvent A and acetonitrile with 0.1% formic acid as solvent B. Two microliters of peptide samples (at a concentration of 0.5 μg μl-1) were loaded on a nanoEase M/Z Symmetry C18 trap column (100 Å, 5 μm; 180 μm × 20 mm; Waters) connected to a nanoEase M/Z HSS T3 column (100 Å, 1.8 μm, 75 μm × 250 mm; Waters) heated to 50 °C. Separation was achieved with a gradient from 2% to 5% B in 2 min, 25% B in 93 min, 35% B in 10 min, and 95% B in 10 min, at a flow rate of 300 nl/min. The mass spectrometer was configured for data-dependent acquisition and operating in top-speed mode with a cycle time of 3 s. Full-scan MS spectra were acquired in the orbitrap analyzer with a mass range of 300 to 1500 m/z and a resolution of 120 k with an automated gain control target value of 4 × 105. Peptides were fragmented using higher energy collisional dissociation (isolation window 1.6 m/z) using a normalized collision energy of 30 with an automated gain control target value of 2e3, and spectra were acquired in the ion trap operated with rapid scan rate. Dynamic exclusion was set to 25 s. Internal lock mass calibration on m/z 371.10124 and 445.12003 was used.
Raw MS files were imported into Progenesis QI (Nonlinear Dynamics, version 4.2.7207.2295) using the High-Mass Accuracy Instrument option. Runs were aligned automatically by Progenesis. From each peptide ion (with default sensitivity in peak picking), a maximum of the top five tandem mass spectra were exported using the charge deconvolution and deisotoping option and a maximum number of 200 peaks per MS/MS. The Mascot generic file (.mgf) was searched using the Mascot Server (Matrix Science, version 2.7.0.1) against a forward and reversed protein sequence database containing the 2609 annotated proteins of M. flagellatus KT (CP000284.1) concatenated with 6721 yeast proteins and 260 known MS contaminants. Parameters for precursor ion tolerance and fragment ion tolerance were set to ±10 ppm and ±0.5 Da, respectively. Trypsin was chosen as protein-cleaving enzyme, and two missed cleavages were allowed. Carbamidomethylation of cysteine was set as fixed modification; oxidation of methionine and carbamylation of the N terminus and lysine as variable modifications. The Mascot search results were imported into Scaffold (Proteome Software, version 5.0.1) using 5% peptide and 10% protein false discovery rate. The Scaffold Spectrum Report was exported and imported back into Progenesis QI. Normalization was performed on all precursor ions. For quantification, all proteins identified with at least two unique peptides were assessed. Proteins were grouped by Progenesis, and the normalized abundance from the most abundant peptide ions (relative quantification using Hi-3) from the same protein group was averaged for each sample. For statistical analysis, one-way ANOVA was applied on the normalized protein abundance. The resulting p values were corrected using the Benjamini–Hochberg correction directly in Progenesis QI. Proteins were considered significantly changing if q value <0.05 and log2 fold change of >2 or <−2. A list of all the identified proteins is available as Supporting Data File S1.
Creation of gene deletion strains
Marker-free gene deletions in M. flagellatus KT were obtained by homologous recombination using the plasmid pK18mobsacB (50). Briefly, ∼700 bp regions upstream and downstream of the gene of interest were amplified by PCR using overlapping primers (Table S4). The two fragments were fused by overlap extension PCR and cloned into the vector using XbaI and HindIII restriction sites, except for Mfla_0908 where XbaI and PstI were used. After transformation into electrocompetent M. flagellatus cells (1 mm cuvette, 1.8 kV, 2.5 h recovery in MMM medium at 37 °C), selection for plasmid integration was performed by plating the cells on MMM agar supplemented with kanamycin (50 μg/ml). To select for the second crossover event, colonies were streaked out on MMM agar supplemented with 5% sucrose. To separate colonies with spontaneous sacB mutations from successful gene deletions, multiple colonies were transferred to MMM agar plates with and without kanamycin. Only colonies that did not grow on the plate with kanamycin were used for further screening by colony PCR and eventually validation by sequencing.
Note that the genome of M. flagellatus KT has been described to contain a large (potentially phage-related) duplication (22), thus both locus tags Mfla_0908 and Mfla_1052 refer to an identical protein sequence (LanP). To verify that lanP was indeed completely absent in our deletion strain, we further genome sequenced this strain and could not detect any reads for lanP.
Analysis of growth in the presence and absence of La3+
To analyze growth in the presence and absence of La3+ (20 μM LaCl3), 20 ml MMM cultures were inoculated in triplicates to a starting absorbance of 0.00002 at 600 nm from independent exponential precultures. Bacterial growth was monitored by measuring the absorbance at 600 nm over time (using a BioPhotometer; Eppendorf). Before each absorbance measurement, samples were vortexed, as clumping was observed in some cases during early growth. For each replicate, the growth rate μ was estimated from an exponential fit to the first 23.5 h of the data, with the initial absorbance set to 2e-5. For each strain, a two-sided t test was used to determine the significance of differences in growth rates between the two conditions (with and without La3+).
RT–qPCR analysis
To analyze Mfla_0344 (xoxF) and Mfla_2038 (mxaA) expression by qPCR in the presence and absence of La3+ (20 μM LaCl3), 20 ml MMM cultures were inoculated in triplicates to a starting absorbance of 0.02 at 600 nm from an overnight preculture. During midexponential growth, cells corresponding to 1 ml of culture with an absorbance at 600 nm of 1 were harvested by centrifugation. For cell lysis, cell pellets were suspended in 1 ml of TRI Reagent (Ambion) and incubated for 5 min at room temperature. For RNA separation from other cell contents, 0.2 ml of chloroform was added followed by vigorous shaking of the tube for 15 s, 3 min of incubation at room temperature, and centrifugation at 12,000g for 15 min at 4 °C. The clear upper phase containing RNA was separated from the other phases, and 0.5 ml of cold isopropanol was added for RNA precipitation. The RNA pellet was washed twice with cold 75% ethanol, dried, and resuspended in 50 μl of RNase-free water (Thermo Fisher Scientific). Complementary DNA (cDNA) was synthesized by applying the High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) using 2 μg of RNA as template, except for culture replicate 3 of the wildtype grown in the presence of LaCl3 for which 1.4 μg were used. The cDNA synthesis incubation steps were 10 min, 25 °C; 120 min, 37 °C; 5 min, 85 °C. For qPCR, samples were diluted 1:20 in 20 μl reaction mixture containing 300 nM primers and FastStart Universal SYBR Green Master Mix (Roche). qPCR was performed in two technical replicates in a QuantStudio 7 Flex (Thermo Fisher Scientific) with standard settings (two-step PCR, 40 cycles, annealing/elongation temperature 60 °C). Mfla_0284 (50s ribosomal protein L22) was selected as housekeeping gene based on the proteomics data. Because of technical issues, only one technical replicate could be considered for the genes Mfla_0284, Mfla_2038, and Mfla_2153 of culture replicate 3 of strain ΔlanP (=ΔMfla_0908) in the presence of LaCl3.
Structure prediction using AlphaFold
For protein structure prediction, the AlphaFold Colab notebook from Deepmind was used (31), and the full LanP (Mfla_0908) sequence was provided as an input. The pLDDT values from AlphaFold were above 90 for all residues except residues close to the termini, thus indicating a confident prediction. The structure was visualized using UCSF ChimeraX (51).
Creation of phylogenetic tree
The sequences for the phylogenetic tree were selected based on an NCBI protein BLAST search (52, 53) against the RefSeq Select proteins (refseq_select) database using the LanP (Mfla_0908) sequence as input. Default search parameters were used, except for the maximal number of target sequences, which was increased to 5000. The resulting list was further filtered by applying a query-coverage cutoff of >85% and an E value cutoff of <1e-20. In addition, sequences that belonged to an unclassified taxonomy were manually removed. Sequence alignment was performed using MUSCLE (54), and the phylogenetic tree was calculated using the IQ-TREE web server with default settings (55). The tree was visualized using the Python framework ETE 3 (56).
Cloning of lanP for expression in E. coli
For N-terminal His10-tagged expression of lanP (Mfla_0908), the gene without the encoded signal peptide (the first 23 amino acids) was amplified by PCR from genomic DNA of M. flagellatus KT using primers that contained NdeI and BamHI restriction sites (Table S4). The PCR product was cloned into the expression vector pET-16b (Novagen) using restriction enzyme cloning. Ligation products were transformed into E. coli DH5α, and correct plasmids were identified by colony PCR and sequencing.
For expression without the His10 tag, the plasmid constructed above was PCR amplified using overlapping primers but omitting the His10 tag (Table S4). Gibson Assembly (NEB) was then used to close the resulting linear plasmid.
For heterologous expression, the plasmids were transformed into E. coli BL21-Gold (DE3) cells (Agilent).
Heterologous expression of lanP
For protein production, LB medium supplemented with ampicillin (100 μg/ml) was inoculated and grown at 37 °C until midexponential phase (absorbance of ∼0.8 at 600 nm). To produce His10-tagged LanP (Mfla_0908), cultures were induced using 0.1 mM IPTG and then switched to 28 °C for overnight expression. To produce untagged LanP, cultures were first cooled down to 16 °C for 30 min, before expression was induced using 1 mM IPTG. The cultures were then incubated at 16 °C overnight, before harvesting by centrifugation.
Purification of His10-tagged LanP
The cell pellet was resuspended using 3 ml of buffer containing 25 mM Tris, 150 mM NaCl, and 20 mM imidazole at pH 7.6. Cell lysis was performed by passing the suspension four times through a French press cell. The resulting lysate was clarified by ultracentrifugation and sterile filtration. Affinity purification was performed using a 1 ml HisTrap HP column (GE Healthcare) and gradient elution from 20 mM to 500 mM of imidazole (in 25 mM Tris, 150 mM NaCl, pH 7.6). Protein-containing fractions were pooled and concentrated using a 10 kDa centrifugal filter (Amicon, Merck). Subsequent size-exclusion chromatography was performed using a Superdex 200 Increase 10/300 GL column (GE Healthcare) and a buffer consisting of 25 mM MOPS and 150 mM NaCl at pH 7.2. The size-exclusion chromatogram and an SDS-PAGE gel of the resulting fractions are shown in Fig. S4.
Purification of untagged LanP
For cell lysis, the pellet was resuspended using 3 to 10 ml of buffer A (20 mM Tris, 20 mM NaCl, pH 5.9). The suspension was then lysed either by passing three times through a French press cell or by sonication (6 mm probe, amplitude 30, 5 s pulses followed by 15 s cooling, 4 min total). The cell lysate was clarified by ultracentrifugation and sterile filtration.
For the purification of untagged LanP (Mfla_0908), multiple chromatographic steps were required to obtain pure protein. During the experiments, the purification was further optimized and improved, resulting in three slightly different protocols.
In the first purification protocol (used for the ICP-MS assays), the lysate was purified using a 1 ml HiTrap Q HP (GE Healthcare) anion-exchange (AEX) column run with a linear gradient of buffer A and buffer B (buffer A with 1 M NaCl). The gradient run at 1 ml/min from 0% B to 50% B in 30 min. The resulting fractions were analyzed by SDS-PAGE, and fractions containing LanP were pooled and concentrated using a 10 kDa centrifugal filter (Amicon, Merck). Next, the sample was purified by size-exclusion chromatography using a Superdex 75 10/300 GL column run with buffer C (25 mM MOPS, 150 mM NaCl, pH 7.2). LanP-containing fractions were pooled, concentrated, and run a second time through the same column to increase purity. A last step of AEX chromatography was then performed using a MonoQ 5/50 GL column (GE Healthcare) and buffers D (20 mM Tris, 20 mM NaCl, pH 7.6) and E (buffer D + 1 M NaCl). A gradient from 0 to 50% B in 30 min at 1 ml/min was applied. Pure fractions were pooled, concentrated, and rebuffered using buffer C in a centrifugal filter.
The second protocol (used for the dye competition assays with Ln3+) started with the same AEX column; however, the following gradient was applied at 1 ml/min: 0% to 20% B in 15 min, 20% B for 7 min, 20% to 35% B in 15 min, and 35% to 100% B in 10 min. Fractions containing LanP were pooled, concentrated, and applied to the MonoQ 5/50 GL column. The same buffers as aforementioned were used (buffers D and E); however, the gradient from 0% to 50% B was elongated to 50 min (at 1 ml/min). The purest fractions were again pooled and concentrated, before loading on the Superdex 75 10/300 GL column and running using buffer C. The size-exclusion chromatogram and the SDS-PAGE gel of the purified protein is shown in Fig. S5.
The third protocol (used for the dye competition assays with nonlanthanide metals and for ITC) was similar to the second protocol; however, slightly modified gradients and buffers were used for the AEX chromatography. Both AEX steps now used the same buffers A (20 mM Tris, 20 mM NaCl, pH 5.9) and B (buffer A with 1 M NaCl). For the first AEX step (HiTrap Q), the gradient was modified as follows: 10% B for 5 min, 10% to 25% B in 15 min, and 25% to 100% B in 10 min. For the second AEX step (Mono Q), the following gradient was used: 10% B for 5 min, 10% to 30% B in 20 min, 30% B for 5 min, and 30% to 100% B in 10 min. The size-exclusion chromatography step remained the same.
All protein concentrations were determined by absorption at 280 nm using a calculated extinction coefficient of 12,950 M−1 cm−1 (Expasy ProtParam).
Competition assays
Competition assays were performed in a total volume of 200 μl in 100 mM Bis–Tris propane buffer at pH 6.5 and containing 10 μM of either AZ3 or XO. About 2 μM of either protein (LanP or BSA) or in case of controls, just protein buffer (25 mM MOPS, 150 mM NaCl, pH 7.2) was added. For the measurement series that investigated the selectivity of LanP for La3+ in the presence of Ca2+, 20 mM CaCl2 was added to the assay. Metal ions (LaCl3, CeCl3, NdCl3, EuCl3, FeSO4, FeCl3, and ZnCl2) dissolved in ultrapure water were titrated in 1 μM steps from a 200 μM stock solution. The binding of ions to the indicator dye was quantified by measuring the absorbance of the solution using a Cary 50 spectrophotometer (Varian) at the following wavelengths: Ln3+, 660 nm (AZ3); Fe2+, 635 nm (AZ3); Fe3+, 615 nm (AZ3); Ln3+, 565 nm (XO); and Zn2+, 530 nm (XO). To prevent any crosscontamination between samples, single-use plastic cuvettes (Uvette, Eppendorf) were used. Absorbance measurements were normalized to the initial absorbance value in the absence of metal. All measurements were performed in triplicates for each condition. The number of binding sites were estimated from the equivalents of Ln3+ required to reach 15% of the maximal absorbance. Linear interpolation was used to approximate the signal between the discrete titration points.
Ultrafiltration-based protein-binding assays
LanP at 4 μM was incubated with three different metal mixes in buffer (25 mM MOPS, 150 mM NaCl, pH 7.2) in a total volume of 300 μl for 30 min at 37 °C. The used metal mixes were 20 μM LaCl3; 100 μM LaCl3, 100 μM CaCl2; 20 μM LaCl3, 20 μM YCl3, 20 μM CeCl3, 10 μM PrCl3, 20 μM NdCl3. To separate protein-bound from free Ln3+, the samples were concentrated in an Amicon Ultra-0.5 ml centrifugal filter with a molecular weight cutoff of 10 kDa (Merck). After centrifugation for 20 min at 12,000g at room temperature, the retentates were washed twice by the addition of 500 μl of buffer followed by centrifugation. The resulting retentates of ∼25 μl were recovered and used for metal quantification by ICP-MS.
ICP-MS analysis of retentates
The retentates obtained from the filtration assay were digested and analyzed on their La, Ca, Y, Ce, Pr, and Nd content by ICP-MS. Concentrated nitric acid (>65% weight; purified by double subboiling distillation) and purified water (≥18.2 MΩ cm; Millipore) were used for sample digestion as well as for dilution of the samples and calibration standards. ICP reference standards (Inorganic Ventures or Merck) were used to prepare internal standard, recovery standard, and calibration standards. Sample preparation was carried out gravimetrically using a Mettler Toledo AT400 balance.
The retentates (approximately 20 mg) were transferred into PTFE digestion vials, and concentrated nitric acid (0.5 ml) and Re recovery standard (0.1 ml of 10 mg l−1 standard) were added. The samples (including blank samples) were digested using a turboWave (MLS GmbH) microwave-assisted system (240 °C, 1200 W at 120 bars for 20 min). The clear and colorless solutions were then transferred into 50 ml centrifuge tubes (TPP) and diluted to 20 ml. In a subsequent dilution step, the samples were further diluted by a factor of 10, and indium stock solution was added as internal standard at a final concentration of 2.5 μg l−1 in 1% HNO3 aqueous solution.
The samples were analyzed using a sector-field inductively coupled plasma mass spectrometer (Element XR, Thermo Fisher) equipped with a micro concentric nebulizer (200 μl min−1, borosilicate glass, glass expansion), a cyclonic spray chamber (borosilicate glass and glass expansion), quartz injector, torch with guard electrode, sampler, and skimmer made of nickel. The instrumental parameters were set to nebulizer gas flow 1 l min−1, plasma gas flow 16 l min−1, auxiliary gas flow 1 l min−1, and ICP power 1330 W. Parameters were optimized for maximum signal intensities and CeO/Ce as well as UO/U oxide ratios of ≤3% and ≤8%, respectively. 89Y, 115In, 139La, 140Ce, 141Pr, 143Nd, 146Nd, 185Re, and 187Re isotopes were measured at low resolution (m/Δm = 300) and 44Ca at medium resolution (m/Δm = 4000) using e-scan mode. Data acquisition was carried out for five runs and five passes resulting in a total measurement time of approximately 2 min per sample.
Six calibration standards containing 0, 0.25, 0.5, 1, 2, and 4 μg l−1 of Y, La, Ce, Pr, and Nd ICP reference standard, 0, 10, 20, 40, 80, 160 μg l−1 of Ca ICP reference standard, and 0, 0.5, 1, 2, 4, 8 μg l−1 of Re ICP reference standard were measured for external calibration. Limits of detection were determined to be 600 pg l−1 (Y), 90 pg l−1 (La), 80 pg l−1 (Ce), 30 pg l−1 (Pr), 170 pg l−1 (Nd), and 10 μg l−1 (Ca). Two retentate samples were further spiked with ICP standards to validate the method and to exclude any matrix effects, resulting in recoveries between 93% and 99%. All samples contained Re as recovery standard, and recoveries between 89% and 101% were determined.
ITC analysis of LanP
ITC measurements were performed using a MicroCal PEAQ-ITC device (Malvern Panalytical). The cell was filled with 50 μM purified LanP in buffer (25 mM MOPS and 150 mM NaCl at pH 7.2). The syringe was filled with 5 mM metal ligand dissolved in the same buffer. For the titration, 30 injections of 0.5 μl with a duration of 1 s and spaced by 150 s were performed (first injection was only 0.2 μl). The reference power was 10 μcal/s, the temperature 25 °C, and the stir speed 750 rpm. For each metal, the heat of dilution was determined from a separate titration of metal into buffer, which was then subtracted (point to point) from the measurement with LanP. All data were analyzed and fitted using the vendor software, before plotting using Python (Python Software Foundation).
Data availability
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD032828.
A list of all the identified proteins is available as Data File S1. Upon request, additional data and material will be made available.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Bernd Roschitzki and Jonas Grossmann from the Functional Genomics Center Zurich for support with the LC–MS/MS measurements. Furthermore, we also acknowledge Laura Brülisauer for help with the proteomics experiment and Bartosz Marcin Lewandowski (Laboratory of Organic Chemistry, ETH Zurich) for assistance with ITC measurements. This work was supported by the Swiss National Science Foundation (grant no.: 310030B_201265).
Author contributions
J. L. H., D. G., and J. A. V. conceptualization; J. L. H., P. K., L. H., T. V., A. M. O., and M. B.-M. Investigation; J. L. H. writing–original draft; P. K. and J. A. V. writing–review & editing; D. G. and J. A. V. supervision.
Reviewed by members of the JBC Editorial Board. Edited by Ruma Banerjee
Footnotes
Present address for Jethro L. Hemmann: Junior Research Group Synthetic Microbiology, Leibniz-Institute for Natural Product Research and Infection Biology, Beutenbergstr. 11a, Jena, 07745, Germany.
Contributor Information
Jethro L. Hemmann, Email: jethro.hemmann@leibniz-hki.de.
Julia A. Vorholt, Email: jvorholt@ethz.ch.
Supporting information
References
- 1.Nakagawa T., Mitsui R., Tani A., Sasa K., Tashiro S., Iwama T., et al. A catalytic role of XoxF1 as La3+-dependent methanol dehydrogenase in Methylobacterium extorquens strain AM1. PLoS One. 2012;7 doi: 10.1371/journal.pone.0050480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daumann L.J. Essential and ubiquitous: the emergence of lanthanide metallobiochemistry. Angew. Chem. Int. Edition. 2019;58:12795–12802. doi: 10.1002/anie.201904090. [DOI] [PubMed] [Google Scholar]
- 3.Cotruvo J.A. The chemistry of lanthanides in biology: recent discoveries, emerging principles, and technological applications. ACS Cent. Sci. 2019;5:1496–1506. doi: 10.1021/acscentsci.9b00642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peplow M. Unlocking the lanthanome. ACS Cent. Sci. 2021;7:1776–1779. doi: 10.1021/acscentsci.1c01357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Featherston E.R., Cotruvo J.A. The biochemistry of lanthanide acquisition, trafficking, and utilization. Biochim. Biophys. Acta Mol. Cell Res. 2021;1868 doi: 10.1016/j.bbamcr.2020.118864. [DOI] [PubMed] [Google Scholar]
- 6.Keltjens J.T., Pol A., Reimann J., Op den Camp H.J.M. PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl. Microbiol. Biotechnol. 2014;98:6163–6183. doi: 10.1007/s00253-014-5766-8. [DOI] [PubMed] [Google Scholar]
- 7.Huang J., Yu Z., Groom J., Cheng J.-F., Tarver A., Yoshikuni Y., et al. Rare earth element alcohol dehydrogenases widely occur among globally distributed, numerically abundant and environmentally important microbes. ISME J. 2019;13:2005–2017. doi: 10.1038/s41396-019-0414-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chistoserdova L., Kalyuzhnaya M.G. Current trends in methylotrophy. Trends Microbiol. 2018;26:703–714. doi: 10.1016/j.tim.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 9.Vu H.N., Subuyuj G.A., Vijayakumar S., Good N.M., Martinez-Gomez N.C., Skovran E. Lanthanide-dependent regulation of methanol oxidation systems in Methylobacterium extorquens AM1 and their contribution to methanol growth. J. Bacteriol. 2016;198:1250–1259. doi: 10.1128/JB.00937-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ochsner A.M., Hemmerle L., Vonderach T., Nüssli R., Bortfeld-Miller M., Hattendorf B., et al. Use of rare-earth elements in the phyllosphere colonizer Methylobacterium extorquens PA1. Mol. Microbiol. 2019;111:1152–1166. doi: 10.1111/mmi.14208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Firsching F.H., Brune S.N. Solubility products of the trivalent rare-earth phosphates. J. Chem. Eng. Data. 1991;36:93–95. [Google Scholar]
- 12.Evans C.H. Springer US; Boston, MA: 1990. Biochemistry of the Lanthanides. [Google Scholar]
- 13.Zytnick A.M., Good N.M., Barber C.C., Phi M.T., Gutenthaler S.M., Zhang W., et al. Identification of a biosynthetic gene cluster encoding a novel lanthanide chelator in Methylorubrum extorquens AM1. bioRxiv. 2022 2022.01.19.476857. [Google Scholar]
- 14.Juma P.O., Fujitani Y., Alessa O., Oyama T., Yurimoto H., Sakai Y., et al. Siderophore for lanthanide and iron uptake for methylotrophy and plant growth promotion in Methylobacterium aquaticum strain 22A. Front. Microbiol. 2022;13:921635. doi: 10.3389/fmicb.2022.921635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groom J.D., Ford S.M., Pesesky M.W., Lidstrom M.E. A mutagenic screen identifies a TonB-dependent receptor required for the lanthanide metal switch in the type I methanotroph “Methylotuvimicrobium buryatense” 5GB1C. J. Bacteriol. 2019;201 doi: 10.1128/JB.00120-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roszczenko-Jasińska P., Vu H.N., Subuyuj G.A., Crisostomo R.V., Cai J., Lien N.F., et al. Gene products and processes contributing to lanthanide homeostasis and methanol metabolism in Methylorubrum extorquens AM1. Sci. Rep. 2020;10:1–15. doi: 10.1038/s41598-020-69401-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cotruvo J.A., Featherston E.R., Mattocks J.A., Ho J.V., Laremore T.N. Lanmodulin: a highly selective lanthanide-binding protein from a lanthanide-utilizing bacterium. J. Am. Chem. Soc. 2018;140:15056–15061. doi: 10.1021/jacs.8b09842. [DOI] [PubMed] [Google Scholar]
- 18.Cook E.C., Featherston E.R., Showalter S.A., Cotruvo J.A. Structural basis for rare earth element recognition by Methylobacterium extorquens lanmodulin. Biochemistry. 2019;58:120–125. doi: 10.1021/acs.biochem.8b01019. [DOI] [PubMed] [Google Scholar]
- 19.Wegner C.-E., Westermann M., Steiniger F., Gorniak L., Budhraja R., Adrian L., et al. Extracellular and intracellular lanthanide accumulation in the methylotrophic Beijerinckiaceae bacterium RH AL1. Appl. Environ. Microbiol. 2021;87 doi: 10.1128/AEM.03144-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chistoserdova L. New pieces to the lanthanide puzzle. Mol. Microbiol. 2019;111:1127–1131. doi: 10.1111/mmi.14210. [DOI] [PubMed] [Google Scholar]
- 21.Wehrmann M., Berthelot C., Billard P., Klebensberger J. Rare earth element (REE)-dependent growth of Pseudomonas putida KT2440 relies on the ABC-transporter PedA1A2BC and is influenced by iron availability. Front. Microbiol. 2019;10:2494. doi: 10.3389/fmicb.2019.02494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chistoserdova L., Lapidus A., Han C., Goodwin L., Saunders L., Brettin T., et al. Genome of Methylobacillus flagellatus, molecular basis for obligate methylotrophy, and polyphyletic origin of methylotrophy. J. Bacteriol. 2007;189:4020–4027. doi: 10.1128/JB.00045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chistoserdova L., Gomelsky L., Vorholt J.A., Gomelsky M., Tsygankov Y.D., Lidstrom M.E. Analysis of two formaldehyde oxidation pathways in Methylobacillusflagellatus KT, a ribulose monophosphate cycle methylotroph. Microbiology. 2000;146:233–238. doi: 10.1099/00221287-146-1-233. [DOI] [PubMed] [Google Scholar]
- 24.Lin H., Deng E.-Z., Ding H., Chen W., Chou K.-C. iPro54-PseKNC: a sequence-based predictor for identifying sigma-54 promoters in prokaryote with pseudo k-tuple nucleotide composition. Nucl. Acids Res. 2014;42:12961–12972. doi: 10.1093/nar/gku1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Long F., Su C.-C., Lei H.-T., Bolla J.R., Do S.V., Yu E.W. Structure and mechanism of the tripartite CusCBA heavy-metal efflux complex. Philosophical Trans. R. Soc. B: Biol. Sci. 2012;367:1047–1058. doi: 10.1098/rstb.2011.0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinchliffe P., Symmons M.F., Hughes C., Koronakis V. Structure and operation of bacterial tripartite pumps. Annu. Rev. Microbiol. 2013;67:221–242. doi: 10.1146/annurev-micro-092412-155718. [DOI] [PubMed] [Google Scholar]
- 27.Yeats C., Rawlings N.D., Bateman A. The PepSY domain: a regulator of peptidase activity in the microbial environment? Trends Biochem. Sci. 2004;29:169–172. doi: 10.1016/j.tibs.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 28.Gallois N., Alpha-Bazin B., Bremond N., Ortet P., Barakat M., Piette L., et al. Discovery and characterization of UipA, a uranium- and iron-binding PepSY protein involved in uranium tolerance by soil bacteria. ISME J. 2021;16:705–716. doi: 10.1038/s41396-021-01113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Josts I., Veith K., Normant V., Schalk I.J., Tidow H. Structural insights into a novel family of integral membrane siderophore reductases. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2101952118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gabler F., Nam S.-Z., Till S., Mirdita M., Steinegger M., Söding J., et al. Protein sequence analysis using the MPI bioinformatics toolkit. Curr. Protoc. Bioinform. 2020;72:e108. doi: 10.1002/cpbi.108. [DOI] [PubMed] [Google Scholar]
- 31.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boratyn G.M., Schäffer A.A., Agarwala R., Altschul S.F., Lipman D.J., Madden T.L. Domain enhanced lookup time accelerated BLAST. Biol. Direct. 2012;7:12. doi: 10.1186/1745-6150-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singer H., Drobot B., Zeymer C., Steudtner R., J. Daumann L. Americium preferred: lanmodulin, a natural lanthanide-binding protein favors an actinide over lanthanides. Chem. Sci. 2021;12:15581–15587. doi: 10.1039/d1sc04827a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deblonde G.J.-P., Mattocks J.A., Wang H., Gale E.M., Kersting A.B., Zavarin M., et al. Characterization of americium and curium complexes with the protein lanmodulin: a potential macromolecular mechanism for actinide mobility in the environment. J. Am. Chem. Soc. 2021;143:15769–15783. doi: 10.1021/jacs.1c07103. [DOI] [PubMed] [Google Scholar]
- 35.Deblonde G.J.-P., Mattocks J.A., Dong Z., Wooddy P.T., Cotruvo J.A., Zavarin M. Capturing an elusive but critical element: natural protein enables actinium chemistry. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abk0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Savvin S.B. Analytical use of arsenazo III: determination of thorium, zirconium, uranium and rare earth elements. Talanta. 1961;8:673–685. [Google Scholar]
- 37.Fernandez-Gavarron F., Brand J.G., Rabinowitz J.L. A simple spectrophotometric assay for micromolar amounts of lanthanum in the presence of calcium and phosphate. J. Bone Mineral Res. 1987;2:421–425. doi: 10.1002/jbmr.5650020509. [DOI] [PubMed] [Google Scholar]
- 38.Basargin N.N., Ivanov V.M., Kuznetsov V.V., Mikhailova A.V. 40 years since the discovery of the arsenazo III reagent. J. Anal. Chem. 2000;55:204–210. [Google Scholar]
- 39.Hogendoorn C., Roszczenko-Jasińska P., Martinez-Gomez N.C., Graaff J.d., Grassl P., Pol A., et al. Facile arsenazo III-based assay for monitoring rare earth element depletion from cultivation media for methanotrophic and methylotrophic bacteria. Appl. Environ. Microbiol. 2018;84 doi: 10.1128/AEM.02887-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rohwer H., Collier N., Hosten E. Spectrophotometric study of arsenazo III and its interactions with lanthanides. Analytica Chim. Acta. 1995;314:219–223. [Google Scholar]
- 41.Macka M., Nesterenko P., Andersson P., Haddad P.R. Separation of uranium(VI) and lanthanides by capillary electrophoresis using on-capillary complexation with arsenazo III. J. Chromatogr. A. 1998;803:279–290. [Google Scholar]
- 42.Lu Y.W., Laurent G., Pereira H. A novel methodology for evaluation of formation constants of complexes: example of lanthanide–arsenazo III complexes. Talanta. 2004;62:959–970. doi: 10.1016/j.talanta.2003.10.030. [DOI] [PubMed] [Google Scholar]
- 43.Loutet S.A., Chan A.C.K., Kobylarz M.J., Verstraete M.M., Pfaffen S., Ye B., et al. In: Trace Metals and Infectious Diseases. Nriagu J.O., Skaar E.P., editors. MIT Press; Cambridge (MA: 2015. The fate of intracellular metal ions in microbes. [PubMed] [Google Scholar]
- 44.Alav I., Kobylka J., Kuth M.S., Pos K.M., Picard M., Blair J.M.A., et al. Structure, assembly, and function of tripartite efflux and type 1 secretion systems in Gram-negative bacteria. Chem. Rev. 2021;121:5479–5596. doi: 10.1021/acs.chemrev.1c00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skovran E., Raghuraman C., Martinez-Gomez N.C. Lanthanides in methylotrophy. Curr. Issues Mol. Biol. 2019;33:101–116. doi: 10.21775/cimb.033.101. [DOI] [PubMed] [Google Scholar]
- 46.Mattocks J.A., Cotruvo J.A. Biological, biomolecular, and bio-inspired strategies for detection, extraction, and separations of lanthanides and actinides. Chem. Soc. Rev. 2020;49:8315–8334. doi: 10.1039/d0cs00653j. [DOI] [PubMed] [Google Scholar]
- 47.Deblonde G.J.-P., Mattocks J.A., Park D.M., Reed D.W., Cotruvo J.A., Jiao Y. Selective and efficient biomacromolecular extraction of rare-earth elements using lanmodulin. Inorg. Chem. 2020;59:11855–11867. doi: 10.1021/acs.inorgchem.0c01303. [DOI] [PubMed] [Google Scholar]
- 48.Hemmerle L., Ochsner A.M., Vonderach T., Hattendorf B., Vorholt J.A. In: Methods in Enzymology. Cotruvo J.A., editor. Academic Press; 2021. Mass spectrometry-based approaches to study lanthanides and lanthanide-dependent proteins in the phyllosphere; pp. 215–236. [DOI] [PubMed] [Google Scholar]
- 49.Ochsner A.M., Christen M., Hemmerle L., Peyraud R., Christen B., Vorholt J.A. Transposon sequencing uncovers an essential regulatory function of phosphoribulokinase for methylotrophy. Curr. Biol. 2017;27:2579–2588.e2576. doi: 10.1016/j.cub.2017.07.025. [DOI] [PubMed] [Google Scholar]
- 50.Schäfer A., Tauch A., Jäger W., Kalinowski J., Thierbach G., Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 51.Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl. Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Altschul S.F., Wootton J.C., Gertz E.M., Agarwala R., Morgulis A., Schäffer A.A., et al. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 2005;272:5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Madeira F., Park Y.m., Lee J., Buso N., Gur T., Madhusoodanan N., et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucl. Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trifinopoulos J., Nguyen L.-T., von Haeseler A., Minh B.Q. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucl. Acids Res. 2016;44:W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huerta-Cepas J., Serra F., Bork P. Ete 3: reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 2016;33:1635–1638. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Teufel F., Almagro Armenteros J.J., Johansen A.R., Gíslason M.H., Pihl S.I., Tsirigos K.D., et al. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022;40:1023–1025. doi: 10.1038/s41587-021-01156-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu S., Wang J., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R., et al. CDD/SPARCLE: the conserved domain database in 2020. Nucl. Acids Res. 2020;48:D265–D268. doi: 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD032828.
A list of all the identified proteins is available as Data File S1. Upon request, additional data and material will be made available.