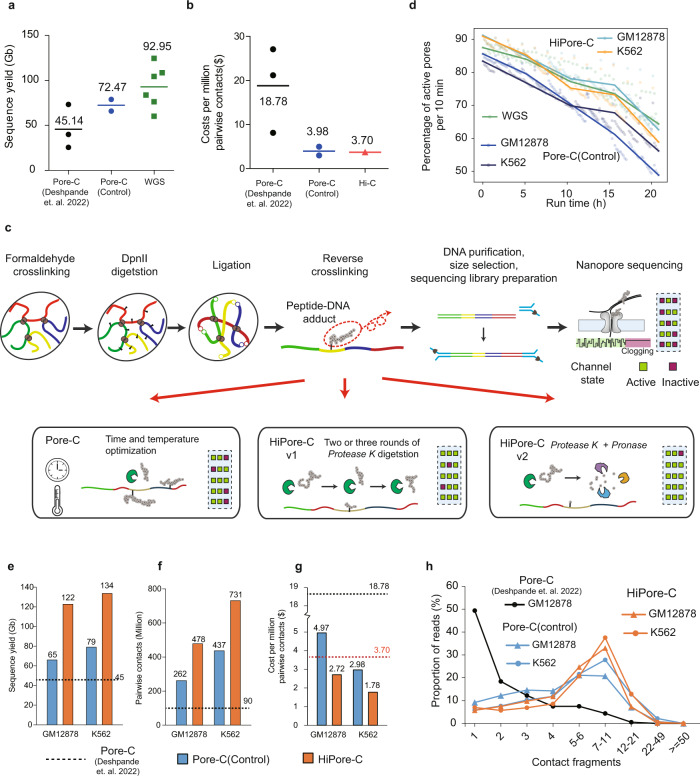
Fig. 1. Solving nanopore clogging increases the output of multiway contact sequencing.
a Comparison of the sequencing yield (Gb) between Pore-C and whole-genome sequencing (WGS) using ONT PromethION flow cells. Datasets of Pore-C (Deshpande et al., 2022)57 and Pore-C (Control) were published and generated in this study, respectively (Pore-C57, n = 3; Pore-C control, n = 2; WGS, n = 6). Lines indicate the mean values. Related to Supplementary Table 1. b Comparison of the costs per million pairwise contacts between Pore-C and Hi-C (Pore-C57, n = 3; Pore-C control, n = 2). The cost of Hi-C is estimated based on the output of the Illumina Nova sequencing platform and the percentage of pairwise contacts that Hi-C can typically produce. Lines indicate the mean values. Related to Supplementary Table 2. c Schematic of the in situ HiPore-C protocols for generating higher-order chromatin interactions. Condition optimization for reverse crosslinking and the effect of nanopore sequencing; Pore-C optimization, bottom left; HiPore-C v1, bottom middle, two or three rounds of reverse crosslinking and protease K digestion; HiPore-C v2, bottom right, reverse crosslinking plus protease K and pronase digestion. Flow cell sequencing channel clogging was compared. Green squares indicate active sequence channels; red squares indicate inactive channels. d Comparison of the decays of the percentage of active pores between WGS, Pore-C (Control), and HiPore-C. X-axis, sequencing time; y-axis, percentage of active pores per 10 min. e-g Comparison of the sequencing yield (Gb), the numbers of virtual pairwise contacts, and the costs between Pore-C and HiPore-C. Related to Supplementary Table 2.h Comparison of the distribution of numbers of ligated fragments in multiway contact long reads between Pore-C and HiPore-C.