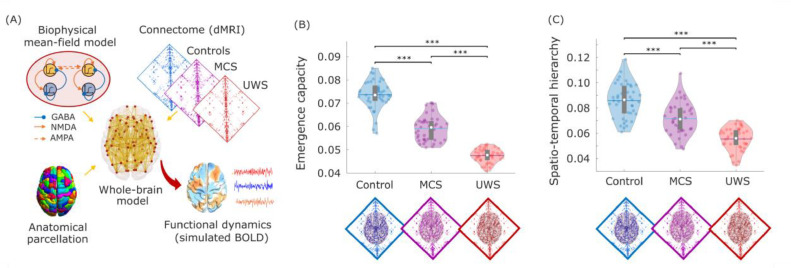
Fig. 5.
Whole-brain models informed by empirical connectomes replicate empirical changes in brain dynamics. (A) Overview of the whole-brain modelling approach to investigate structure-function relationships. The whole-brain model is based on local biophysical models of excitatory and inhibitory neuronal populations, corresponding to brain regions as defined by an anatomical parcellation, interconnected by a network of structural connections obtained from diffusion MRI from each group of subjects (healthy controls, MCS and UWS patients). The whole-brain model has one free parameter, the global coupling G, which is selected as the value just before the simulated firing rate becomes unstable. (B) Emergence capacity is highest in the dynamics simulated from control connectome, in line with empirical results. (C) Spatio-temporal hierarchical character is highest in the dynamics simulated from control connectome, in line with empirical results. Each data-point corresponds to one of 40 simulations obtained from each whole-brain model. White circle, median; centre line, mean; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range. *** p < 0.001, FDR-corrected.