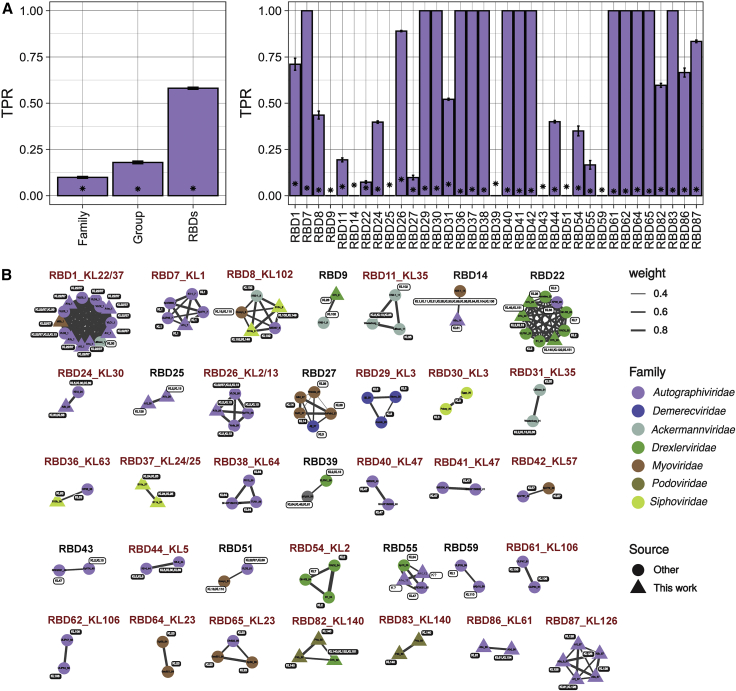
Figure 4.
RBD-based prediction of phage capsular tropism
(A) Left panel: sensitivity or TPR of different phage traits to predict infections in capsular types of bacteria. Three independent calculations were performed. For RBDs, the average of the 35 RBD clusters across calculations is shown. Asterisks represent the mean TPR values obtained with randomized data (null expectation). Right panel: TPR for each individual RBD cluster. Bars represent the standard error of independent calculations (n = 3).
(B) Representation of the 35 RBD similarity clusters obtained. RBDs are named by the phage name followed by the phage protein in which they were detected. Clusters labeled in red are those for which a consensus capsular tropism could be obtained. Colors of nodes indicate phage taxonomic families. Sequences obtained in the present work are shown as triangles and previous published sequences as circles. Edge weight (width) represents amino acid identity between RBDs within the same cluster. Each RBD is annotated with the corresponding phage CLT tropism. White labels represent tropisms that did not match the consensus CLT of an RBD cluster.
See also Table S4.