Figure 5.
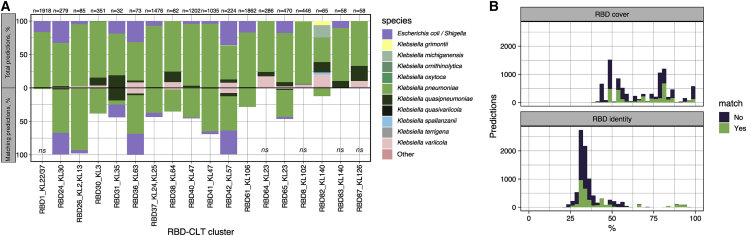
Validation of RBD-based capsular tropism prediction using prophage sequences
(A) Ability of the consensus RBD clusters to predict the CLT tropism of prophages obtained from the RefSeq database as described in the main text. The percentage of total and matching predictions by bacterial species is shown. The n values indicate the total predictions for each RBD. “Other” refers to spp. within Klebsiella sp. and also from other genera. Ns denotes no significance by Fisher’s exact test (p > 0.05).
(B) Distribution of RBD coverage and identity for prophage predictions. Colors indicate whether the CLT of the lysogenized host matched the consensus CLT of the RBD cluster used as a query. Hits with ≤40% RBD cover were not considered.