Abstract
During mammalian spermiogenesis, major restructuring of chromatin takes place. In the mouse, the histones are replaced by the transition proteins, TP1 and TP2, which are in turn replaced by the protamines, P1 and P2. To investigate the role of TP2, we generated mice with a targeted deletion of its gene, Tnp2. Spermatogenesis in Tnp2 null mice was almost normal, with testis weights and epididymal sperm counts being unaffected. The only abnormality in testicular histology was a slight increase of sperm retention in stage IX to XI tubules. Epididymal sperm from Tnp2-null mice showed an increase in abnormal tail, but not head, morphology. The mice were fertile but produced small litters. In step 12 to 16 spermatid nuclei from Tnp2-null mice, there was normal displacement of histones, a compensatory translationally regulated increase in TP1 levels, and elevated levels of precursor and partially processed forms of P2. Electron microscopy revealed abnormal focal condensations of chromatin in step 11 to 13 spermatids and progressive chromatin condensation in later spermatids, but condensation was still incomplete in epididymal sperm. Compared to that of the wild type, the sperm chromatin of these mutants was more accessible to intercalating dyes and more susceptible to acid denaturation, which is believed to indicate DNA strand breaks. We conclude that TP2 is not a critical factor for shaping of the sperm nucleus, histone displacement, initiation of chromatin condensation, binding of protamines to DNA, or fertility but that it is necessary for maintaining the normal processing of P2 and, consequently, the completion of chromatin condensation.
The development of spermatids into spermatozoa, termed spermiogenesis, is characterized by striking morphological and molecular transformations (43). In elongating and condensing spermatids, major restructuring of the somatic chromatin takes place in which the histones are first replaced by a group of arginine- and lysine-rich proteins called transition proteins (TPs), which are in turn replaced by protamines (42). At that time, transcription ceases, the nucleosomal-type chromatin is transformed into a smooth fiber, and condensation begins (30).
TP1 and TP2 are the predominant TPs found in rodent spermatids (24). TP1 is a 6.2-kDa, highly basic chromosomal protein with evenly distributed basic residues (32, 34). TP2, by contrast, is a 13-kDa protein with distinct structural domains (39). The carboxyl third of the molecule is enriched in basic residues and is likely to be a major site of electrostatic DNA binding (14), whereas the amino-terminal region has two proposed zinc fingers (41). The preferential binding activity of TP2 to CpG sequences, which are often associated with promoter regions, is dependent on zinc (36).
TP2, in contrast to the highly conserved TP1 (34), is poorly conserved across mammals, showing only 75% nucleotide and 50% amino acid homologies (1, 29). Whereas TP1 is abundantly expressed in all mammals studied (27), representing 60% of the basic proteins of mouse step 12 to 13 spermatid nuclei (62), TP2 levels vary (1). For example, the mRNA in human testis is detectable only by reverse transcription-PCR (54), but in mice the protein constitutes about 30% of the basic proteins of step 12 to 13 spermatid nuclei (62). Based on these considerations, TP1 should have the more significant role in mouse sperm development, unless TP2 has a specific role that cannot be complemented by another protein.
The timing of the appearance of the TPs has been most extensively studied in the rat. Some studies indicated that both proteins are present at significant levels only in the condensing step 13 to 15 spermatids (1, 24, 27, 44). However, others suggested that TP2 first appears at step 10 or 11 during nuclear elongation, whereas TP1 appears slightly later at step 11 or 12 (31, 46). In the mouse, initial studies indicated that TP2 first appears at step 12, reaches a maximum at step 13, but disappears during step 14 (1), but a recent study reported the presence of TP2 as early as step 10 (60).
In the mouse, the TPs are replaced by two protamines, P1 and P2 (8). Whereas P1 is synthesized as the mature protein, P2 is synthesized as a precursor (pre-P2) with 106 residues (61) and is sequentially cleaved to produce six identifiable intermediates (int-P2) and the mature form with 63 residues (15). The genomic locus for TP2 is found in the same gene cluster as the protamines in all mammals examined (53), suggesting that they have an evolutionary relationship and therefore may share some common functions.
Different functions have been suggested for the TPs, including nuclear shaping, histone removal, transcriptional repression, chromatin condensation, and, most recently, repair of the DNA strand breaks that normally transiently occur during the removal of the nucleosomes (12). In vitro, both TP1 and TP2 stabilize DNA in a nonsupercoiled state and bring DNA molecules in close proximity (38). Other studies indicated that the two TPs have distinct roles. TP1 has been reported to decrease the melting temperature of DNA and decrease nucleosome compaction (55, 56), whereas TP2 does the opposite (7), leading to the suggestion that TP1 is involved in histone removal and TP2 is involved in chromatin condensation. Finally, the preferential binding of TP2 to CpG islands may indicate a role for it in global repression of transcription (41).
To test these predictions and to understand the exact role of TPs in spermiogenesis, we first generated Tnp1-null mice (62). Although we found that TP1 was not essential for histone displacement or chromatin condensation, probably because its absence may be partially compensated for by TP2, the absence of TP1 did result in an abnormal pattern of chromatin condensation and in reduced fertility. Subsequently, we generated mice lacking TP2 by targeted deletion of the Tnp2 gene. In this paper, we describe the effects of this mutation on the appearance and protein composition of various stages of spermatids and on chromatin structure and function of spermatozoa.
MATERIALS AND METHODS
Construction of the Tnp2-targeting vector.
A Stratagene 129 mouse genomic library in the λ-FixII vector was screened with a Tnp2 cDNA (33). The restriction sites of one of the positive clones overlapped with a sequenced protamine gene cluster clone (GenBank accession number Z47352) for 10.5 kb (53), which was used to select additional restriction sites used in constructing the replacement vector from our clone (Fig. 1A).
FIG. 1.
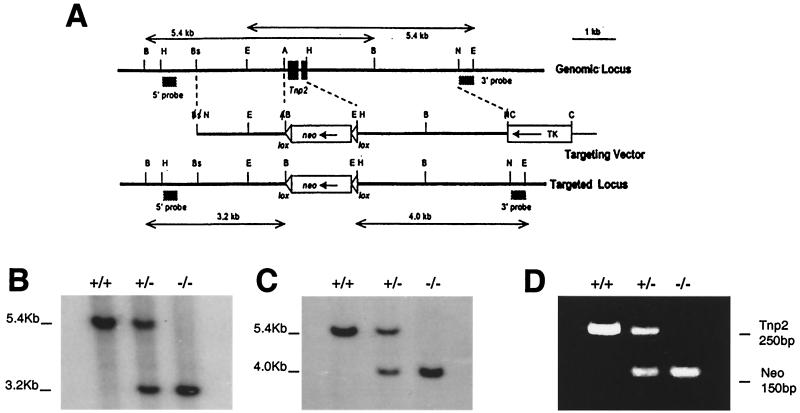
Targeted deletion of the Tnp2 gene. (A) Genomic Tnp2 locus, the replacement vector used, and the targeted locus. Restriction sites: A, AvaI; B, BamHI; Bs, BsmI; C, ClaI; E, EcoRI; H, HindIII; N, NotI. The positions of the probes used to distinguish the targeted locus from the endogenous locus are indicated. (B and C) Southern blot genotyping performed by probing BamHI digests with the 5′ probe (B) or probing EcoRI digests with the 3′ probe (C). (D) PCR genotyping using primers for Tnp2 and neo.
A Bluescript KS plasmid containing a PGKneobpA(lox) cassette was used for construction of the Tnp2-targeting vector (Fig. 1A). First, the 5′ and 3′ flanking regions of the Tnp2 gene were individually cloned into Bluescript SK using the HindIII and EcoRI sites, respectively. To clone the 5′ region of homology into the KS plasmid, the BsmI (nucleotide [nt] 6989) and AvaI (nt 8769) sites were modified by adding NotI and BamHI linkers, respectively, and then this region was inserted into the NotI and BamHI sites of the Bluescript KS plasmid. Next the ClaI site in the Bluescript SK plasmid containing the 3′ region was eliminated and the NotI site (nt 13031) at the end of the 3′ region was converted to a ClaI site. The 3′ region of homology was excised with HindIII (nt 9431) and ClaI and was inserted into those restriction sites in the KS plasmid, and then a ClaI fragment carrying the thymidine kinase gene cassette (28) was inserted into this ClaI site.
Generation of Tnp2 mutant mice.
The NotI-linearized Tnp2-targeting vector was electroporated into AB-1 ES cells, which subsequently were cultured in the presence of G418 and 1-(2-deoxy-2-fluoro-β-d-arabinofuranosyl)-s-iodouracil (FIAU) on mitotically inactivated STO fibroblasts (28). Resistant ES cell clones were screened by Southern blotting. The 5′ probe external to the region of vector homology was a 350-bp EcoRI-BamHI fragment from a P2 (Prm2) cDNA clone (26), and the 3′ probe was a NotI-EcoRI fragment (nt 13031 to 13405) external to the 3′ region of homology. Correctly targeted ES clones were identified by Southern blotting and were microinjected into C57BL/6J (B6) blastocysts, which were transferred to pseudopregnant foster mothers.
Male chimeras from the ES cell clones were bred first with B6 females. The chimeras that produced high percentages of offspring with agouti fur were then bred with 129/SvEv females to obtain inbred mice. Experiments were performed with the mice derived from clone 2C8 on the 129 background except where noted. Heterozygous offspring for the Tnp2 null mutation were identified by Southern blotting of tail DNA using external probes or by PCR using primers to amplify a portion of the neo gene. Mice produced by mating two Tnp2 heterozygotes were used in all experiments. The mice were genotyped with primers for amplification of both neo (21) and Tnp2 (upstream, 5′-CAG AGC CTT CCC ACC ACT CAT-3′; downstream, 5′-CCC TTC AAA GGT CTT CCT GTT-3′).
To remove neo from the Tnp2 locus, a Tnp2−/− female with the loxP-flanked neo insert was bred with a homozygous CMV-Cre transgenic male on a B6 background (2). Tnp2+/− offspring carrying the recombined Tnp2-null locus with neo deleted were bred with wild-type 129 mice to segregate the Cre transgene and confirm germ line transmission of the deletion of neo. Tnp2+/− Cre− offspring were intercrossed to generate the different genotypes, which were identified by PCR with primers in the Tnp2 5′ homology region (5′-ACT TTC CAG GCC AAG CCA CAG G-3′) and the 3′ homology region (5′-CCG GGC GGT TAA AAG CAC TGA C-3′), as well as with the internal Tnp2 primers described above.
Isolation of spermatogenic cells and preparation of nuclei.
For isolation of sonication-resistant spermatid nuclei (SRS), which represent step 12 to 16 spermatids, our standard procedures (59, 62) were modified as follows. The concentration of phenylmethylsulfonyl fluoride (PMSF) was increased to 0.5 mM before homogenization and a protease inhibitor cocktail (standard cocktail. [1 mM phenylmethylsulfonyl fluoride, 1 μg p-aminobenzamidine per ml, 1 μg of leupeptin per ml, 1 μg of pepstatin A per ml] unless otherwise mentioned) was added to the buffer at each later step in the procedure. After sonication for 3 min at 50% of maximum power using a 3-mm-diameter probe (model 250 Sonifier, Branson Ultrasonics, Danbury, Conn.), each milliliter of spermatid nuclei was mixed with 9 ml of 83.5% (wt/vol) sucrose and centrifuged through a 7-ml cushion of 83.5% (wt/vol) sucrose at 100,000 × g in a Beckman SW 27 rotor for 1 h. The pellet was washed with MP buffer (5 mM MgCl2, 5 mM sodium phosphate, pH 6.5).
Spermatozoa were isolated for protein analysis by mincing the cauda epididymis in Dulbecco's phosphate-buffered saline (PBS). The released sperm were filtered through an 80-μm screen and pelleted. After resuspension in water containing protease inhibitors, the epididymal sperm were sonicated as done for SRS, and the nuclei were purified by centrifugation through 5% sucrose in a 0.2× concentration of MP buffer with 0.25% Triton X-100 at 740 × g for 5 min. The pellet was washed with MP buffer.
Preparation and analysis of nuclear proteins.
All extraction and preparation procedures were performed at 4°C. For selective extraction of TP1, TP2, and histone H1, two testes first were homogenized in water with protease inhibitors and sonicated for 1 min in a 51-mm-diameter cup-horn sonicator. Basic proteins were then extracted with 0.25 M HCl and precipitated overnight with 3.5% trichloroacetic acid (TCA), and the supernatant was retained (11). In all procedures, the proteins were finally precipitated with 25% TCA and the precipitates were washed with acidified acetone, followed by acetone, and dried (47).
For extraction of basic proteins from the SRS, nuclei were incubated in 200 μl of water with protease inhibitors and 10 mM dithiothreitol (DTT) for 30 min and then extracted with 0.5 M HCl (62).
The epididymal sperm nuclear proteins were prepared by disrupting the nuclei with guanidine hydrochloride and DTT and dissociating the proteins with urea, mercaptoethanol, and NaCl (4). The DNA was precipitated by addition of HCl to 0.5 M and removed by centrifugation. The solutes were removed by dialysis against 0.01 M HCl–10 mM DTT, and proteins were precipitated. Parts of these protein preparations were aminoethylated by incubation with ethyleneimine (6) to allow separation of P1 and the mature form of P2.
Proteins were separated by electrophoresis in acid-urea–18% polyacrylamide gels. Several different protein amounts were loaded for each sample to determine the linear range for Coomassie blue-stained bands. Gels were scanned with a Molecular Dynamics laser-scanning densitometer, and protein was quantified by using IMAGEQUANT version 5.0 software (Molecular Dynamics, Sunnyvale, Calif.).
Immunoblot analysis.
The nuclear proteins were electroblotted from the gels onto a Hybond polyvinylidene difluoride membrane (Amersham Pharmacia Biotech, Piscataway, N.J.) in 0.7% acetic acid at 320 mA for 20 min. After staining with Ponceau S, strips were cut off based on the position of marker extracts and blocked with 5% nonfat dry milk in PBS with 0.1% Tween 20 (PBS-T). The strips were incubated for 1 h at 25°C with anti-TP1 antiserum (1:6,000), anti-TP2 antiserum (1:1,500) (both courtesy of Stephen Kistler, University of South Carolina, Columbia), and anti-H1 antiserum raised against calf thymus H1 (1:1,000) (courtesy of Sylviane Muller, Institut de Biologie Moleculaire et Cellulaire, Strasbourg, France) diluted in PBS-T. After being washed three times for 15 min each in PBS-T, the blots were incubated with anti-rabbit immunoglobulin G antibody linked to horseradish peroxidase. The binding to the proteins was detected with ECL-plus reagents (Amersham Pharmacia), imaged with a Molecular Dynamics Storm gel and blot system, and quantified using IMAGEQUANT software.
RNA isolation and Northern blot analysis.
Total RNA was extracted from mature mouse testes with the RNAgents Total RNA Isolation System (Promega, Madison Wis.). Plasmids containing Tnp1 and Tnp2 cDNAs were provided by Kenneth Kleene (University of Massachusetts, Boston) (32, 33). Prm1 and Prm2 cDNA plasmids (61) were purchased from the American Type Culture Collection. Northern blotting and hybridization were performed as described elsewhere (52). The 400-bp Tnp1, 550-bp Tnp2, 480-bp Prm1, and 550-bp Prm2 fragments were used as probes, which hybridized with bands centered at 600, 700, 550, and 760 bp, respectively, on the blot. A 0.7-kb fragment from a plasmid containing rat cyclophilin cDNA, provided by Miles Wilkinson (M.D. Anderson Cancer Center, Houston, Tex.), was used as a control probe and hybridized with a 720-bp band (58). The levels of mRNA were quantified using IMAGEQUANT software.
Light and electron microscopy.
For light microscopy, testes were fixed overnight in Bouin's solution and embedded in paraffin. Tissue sections, 4 μm thick, were stained with periodic acid-Schiff stain and counterstained with hematoxylin. For analysis of sperm retention, averages of 34, 7, 9, and 19 round or nearly round stage VIII, IX, X, and XI tubules, respectively, were counted in three mice of each genotype.
For preparation of samples for electron microscopy, mice were given heparin (1.3 IU/g of body weight) intraperitoneally 15 min prior to anesthesia, and the testes and epididymides were cleared by cardiac perfusion with saline and then perfusion fixed in vivo with buffered glutaraldehyde (57). Tissues were washed in buffer (three times overnight), postfixed in an osmium-ferrocyanide mixture for 1 h (49), dehydrated, infiltrated and embedded with Araldite, and examined by light microscopy. Selected areas were chosen, based on light microscopy of 0.5 to 1.0-μm-thick sections stained with toluidine blue, and thin-sectioned, and sections displaying silver-gold interference colors were used.
Testicular and epididymal sperm counts.
Individual testes were homogenized in 1 ml of deionized water for 5 min using a Polytron homogenizer at setting 9 and sonicated in a 51-mm-diameter cup-horn sonicator for 3 min to remove sonication-sensitive cells. Each cauda epididymis was minced in 1 ml of PBS, and after 30 min the tissue pieces were separated from sperm by pipetting and then passing through an 80-μm-pore-size filter. All counts were performed using a hemacytometer.
Sperm morphology.
Air-dried smears were prepared from sperm in PBS, stained with hematoxylin, and examined using light microscopy at a magnification of ×1,000. Head or tail morphology was determined independently for each mouse with separate counts of at least 100 cells each.
Preparation of nuclei for flow cytometry.
The epididymides from each mouse were minced in 2 ml of TNE buffer (0.1 M Tris, 0.15 M NaCl, and 1 mM EDTA, pH 7.4). After ∼30 min at room temperature, the suspension was pipetted several times and the debris was removed by filtering through 80-μm mesh. Sperm were diluted to 1 × 106 to 2 × 106 cells/ml. Because it proved necessary to use isolated nuclei, sperm heads were separated from tails in two ways, both yielding >90% separation.
Parts of the samples were treated with trypsin (code TRL; Worthington Biochemical Co., Lakewood, N.J.) at 0.05 mg/ml for 30 min at room temperature, followed by addition of soybean trypsin inhibitor (Worthington Biochemical Co.) to 0.5 mg/ml. This procedure breaks the tails off from the heads but leaves the sperm tails intact and has been reported not to cause any apparent damage to sperm head structures (45).
Another aliquot was subjected to mild sonication at about 4°C for 30 s, with an output power setting of 70% of maximum in a 51-mm-diameter cup-horn sonicator (W185; Branson Sonic Power Co.). Sperm heads remained intact, but the tails were fragmented into pieces. Sonication does not alter the acridine orange (AO) staining properties of sperm (20).
Propidium iodide staining and flow cytometry.
Aliqouts of sperm nuclei were stained with 25 μg of propidium iodide (Sigma) per ml in the presence of 0.3% Nonidet P-40 (Sigma) and RNase (40 μg/ml) (Boehringer Mannheim) for 1 h at room temperature (35). Samples were stored overnight at 4°C before being analyzed with an Epics 752 flow cytometer (Beckman-Coulter Corp., Hialeah, Fa.). The excitation wavelength was 488 nm, and the propidium iodide fluorescence signals (area under peak) were collected in list mode using a 610-nm long-pass filter. For normalization of the data, samples from a wild-type mouse were analyzed both before and after analysis of samples from all mutation-carrying mice.
AO staining and flow cytometry.
Other aliquots of sperm nuclei were stained with AO, a metachromatic dye that produces green fluorescence when bound to double-stranded DNA and red fluorescence when bound to single-stranded DNA. A 0.2-ml aliquot was mixed with 0.4 ml of acid denaturation solution (0.15 M NaCl, 0.08 N HCl, and 0.1% Triton X-100, pH 1.4) (18). After 30 s, the cells were stained with 1.2 ml of AO solution (6 μg/ml), (Polysciences, Warrington, Pa.) in a buffer consisting of 0.1 M citric acid, 0.2 M Na2HPO4, 1 mM EDTA, and 0.15 M NaCl (pH 6.0).
Samples were measured in a FACScan flow cytometer (Becton Dickinson, Mountain View, Calif.) with a 488-nm excitation wavelength and a 20-μm beam width, starting 3 min after staining. The green and red fluorescence signals were collected using 530-nm band-pass and 650-nm long-pass filters, respectively. The signals were stored in list mode and analyzed using WinList (Verity Software House, Topsham, Maine).
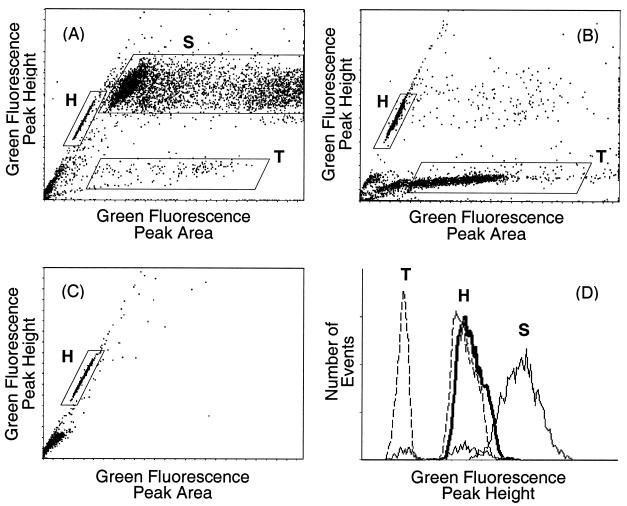
Bivariate histograms obtained with acid-treated, initially intact epididymal sperm from wild-type mice revealed a complex pattern (Fig. 2A) that had not been described previously (18). The acid treatment dissociated some of the sperm heads from the tails (45), producing a mixture of intact sperm, free sperm heads, and free sperm tails. Based on the relative fluorescence peak heights and widths and the changes in subpopulations with different preparative techniques (Fig. 2B and C), the regions marked S, T, and H in Fig. 2 must represent intact sperm, free sperm tails, and heads, respectively. The median green fluorescence intensity of the sperm tails was about 40% of the intensity of the heads, corresponding to microscopic observation of faint staining along the length of the tail (Fig. 2D). Hence, it was essential to use isolated sperm heads if nuclear changes were to be accurately measured in this study. The fluorescence from sperm heads was measured after gating specifically on the head cluster in the scattergrams of green peak height versus green peak width.
FIG. 2.
Bivariate histograms of green fluorescence peak height versus peak area for acid-denatured, AO-stained preparations from wild-type mice. Regions containing intact sperm (S), free sperm heads (H), and free sperm tails (T) are marked. (A) Intact sperm. (B) Trypsin-treated sperm. (C) Sonicated sperm. (D) Histograms of green fluorescence peak height for the indicated gated areas from intact sperm (thin solid lines), trypsin-treated sperm (dashed lines), and sonicated sperm (thick solid line).
The relative amount of red fluorescence was calculated by dividing the red fluorescence by the total (red plus green) fluorescence. This parameter, called αt (18), represents the amount of denatured, single-stranded DNA over the total cellular DNA.
Statistical analysis.
Student's t test was used to compare averages in different experimental groups.
RESULTS
Targeted deletion of Tnp2 in the mouse germ line.
The Tnp2 gene was disrupted in ES cells by homologous recombination with a targeting vector in which the entire Tnp2 gene was replaced by a neo gene flanked by loxP sites (Fig. 1A). Germ line transmissions were obtained from chimeras of two independent clones. Clone 2C8 was selected for detailed analysis; clone 2E3 was used for comparison with 2C8. Heterozygous offspring produced by each chimera were intercrossed. Their offspring were genotyped by Southern blotting of tail DNA using external probes (Fig. 1B and C) or by PCR (Fig. 1D), which was in agreement with Southern analysis. Out of 206 offspring, there were 50 wild-type, 111 heterozygous, and 45 homozygous mice, a ratio consistent with 1:2:1 Mendelian inheritance.
Heterozygous (Tnp2+/−) and homozygous (Tnp2−/−) mutant mice were normal in appearance, growth, and health. Their body weights and seminal vesicle weights (Table 1) were comparable to those of their wild-type littermates, indicating normal development or androgen levels.
TABLE 1.
Organ weights and numbers of sperm in Tnp2 mutant micea
| Tnp2 genotype | Seminal vesicle wt (mg) | Testis wt without tunica (mg) | Sperm heads per testis (106) | Sperm per cauda epididymis (106) |
|---|---|---|---|---|
| Tnp2+/+ | 343 ± 25 | 105 ± 4 | 25 ± 2 | 13 ± 1 |
| Tnp2+/− | 319 ± 21 | 108 ± 5 | 22 ± 2 | 14 ± 1 |
| Tnp2−/− | 278 ± 33 | 107 ± 3 | 23 ± 2 | 11 ± 3 |
Mice (n = 10) were used at 24 weeks of age. All values are means and standard errors.
Northern blotting showed that the levels of Tnp2 mRNA in heterozygous mice were about 50% of those in wild-type mice and undetectable in Tnp2-null mice (Table 2). Although Prml and Prm2 mRNA levels appeared to be slightly but significantly reduced when cyclophilin was used for comparison, there was no significant reduction when Tnp1 mRNA, which is in the same cell type as the protamines and whose message level should not be affected by the mutation in Tnp2, was used for normalization. Western blotting of 3.5% TCA-soluble proteins of total testis confirmed that TP2 protein was absent in Tnp2−/− mice (Fig. 3B).
TABLE 2.
Levels of mRNA for late spermatid basic nuclear proteins in Tnp2 mutant mice
| neo | Tnp2 genotype | Ratio of mRNA level to that in the wild typea
|
|||
|---|---|---|---|---|---|
| Tnp1 | Tnp2 | Prm1 | Prm2 | ||
| Presentb | Tnp2+/− | 0.97 ± 0.07 | 0.44 ± 0.02 | 0.93 ± 0.07 | 0.87 ± 0.06 |
| Tnp2−/− | 0.93 ± 0.06 | 0 | 0.84 ± 0.05* | 0.83 ± 0.03* | |
| Absentc | Tnp2+/− | 0.99 ± 0.06 | 1.06 ± 0.05 | 1.02 ± 0.02 | |
| Tnp2−/− | 1.10 ± 0.07 | 1.19 ± 0.02* | 1.08 ± 0.10 | ||
All values (means and standard errors, n = 3 mice) were first normalized to levels of cyclophilin mRNA in the same lane. *, Ratio significantly different from 1 (P < 0.05), but the ratio was not significantly different from 1 when Tnp1 mRNA was used for normalization.
neo was present in the Tnp2-null alleles; 129 genetic background.
neo was removed from Tnp2-null alleles by Cre recombinase; hybrid genetic background.
FIG. 3.
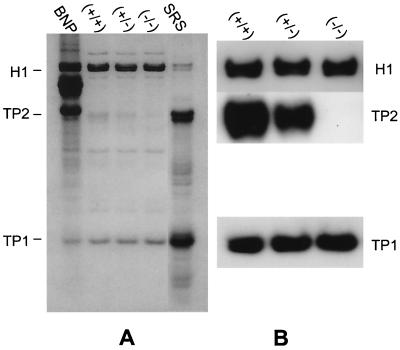
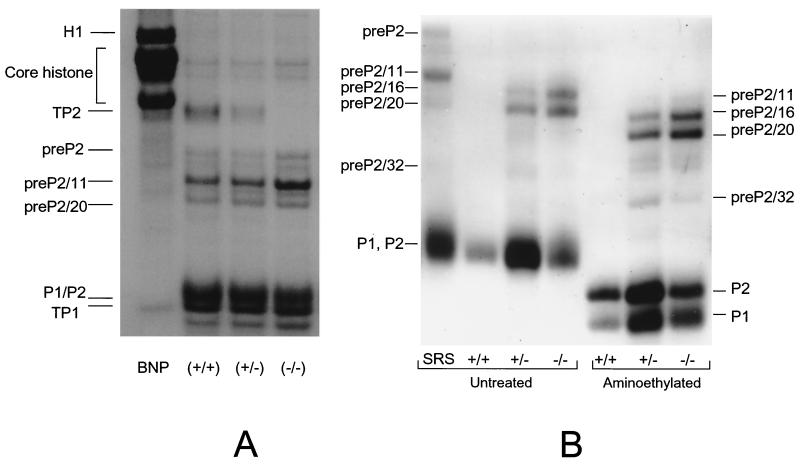
Analysis of 3.5% TCA-soluble proteins from testes of wild-type, heterozygous, and Tnp2 mutant mice by acid-urea gel electrophoresis. (A) Coomassie blue-stained gel. A total basic nuclear protein extract (BNP) from unfractionated wild-type mouse testis nuclei was used as a marker. (B) Immunoblots using antibodies to TP1, TP2, and H1.
Subtly abnormal spermatogenesis in the absence of the Tnp2 gene.
When Tnp2 mutant mice were mated with individual 8-week-old B6 females, the percentage of mutant mice that were fertile was not significantly different from that of normal mice (Table 3). However, the average litter size produced by the Tnp2-null males (3.9 ± 0.7) was significantly reduced from that observed for the wild type (7.4 ± 0.4).
TABLE 3.
Fecundity and sperm morphology of Tnp2 mutant mice
| Genetic background | Tnp2 genotype | Fecundity of Tnp2 mutant males during an 8-week mating period
|
% of sperm (mean ± SEM) witha:
|
||||
|---|---|---|---|---|---|---|---|
| No. fertile/total | Litters per fertile pair (mean ± SEM) | Litter size (mean ± SEM)b | Normal tail morphologyb | Normal head morphology | Head with blunt tips | ||
| 129 | Tnp2+/+ | 10/10 | 1.7 ± 0.2 | 7.4 ± 0.4 | 85.7 ± 3.0 | 94.3 ± 0.3 | 2.7 ± 0.3 |
| Tnp2+/− | 10/10 | 2.1 ± 0.1 | 7.6 ± 0.4 | 81.7 ± 1.4 | 95.7 ± 0.4 | 2.0 ± 0.4 | |
| Tnp2−/− | 8/9 | 2.3 ± 0.2 | 3.9 ± 0.7* | 40.5 ± 1.2* | 94.5 ± 0.7 | 2.1 ± 0.6 | |
| 129/B6 | Tnp2+/+ | 10/10 | 2.6 ± 0.2 | 6.6 ± 0.4 | 91.0 ± 2.7 | 98.5 ± 0.5 | 1.3 ± 0.5 |
| Tnp2+/− | 85.6 ± 5.8 | 98.1 ± 0.8 | 1.8 ± 0.8 | ||||
| Tnp2−/− | 4/5 | 2.0 ± 0.8 | 4.5 ± 1.7 | 70.8 ± 10.4 | 92.1 ± 3.6 | 5.5 ± 2.3 | |
Between four and seven mice were analyzed for each genotype.
*, significantly different from results for both Tnp2+/+ and Tnp2+/− mice (P < 0.05)
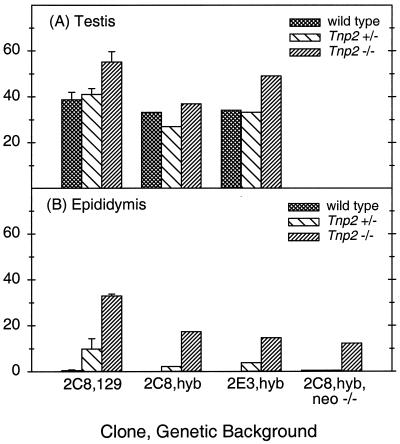
Testis weights and testicular sperm counts were normal in the Tnp2−/− mice, suggesting complete spermatogenesis (Table 1). Furthermore, the epididymal sperm counts in Tnp2−/− mice were unchanged, indicating that most of the sperm were indeed released from the testis.
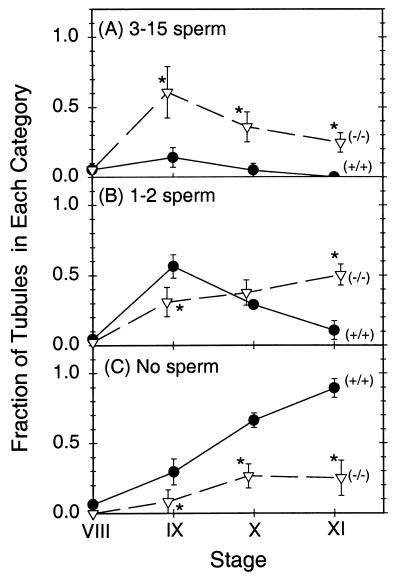
Light microscopic examination showed generally normal testicular histology in Tnp2−/− mice. Most of the mature spermatids, referred to as sperm, were released at the appropriate stage, as 90% of stage VIII but none of the stage IX tubules contained numerous (>15) sperm, just as in the wild type. However, a more quantitative analysis revealed retention of a few sperm (Fig. 4). Whereas in normal mice only 14% of stage IX tubules retained 3 to 15 sperm, 60% of the tubules in Tnp2-null mice showed this level of retention (Fig. 4A). Significant increases in sperm retention were also observed at stages X and XI. Tnp2 heterozygotes showed significant increases in sperm retention in stages IX and X, similar to the Tnp2-null mice, but not in stage XI, which appeared to be similar to the wild type. The sperm retention in the Tnp2 mutant mice is attributed to subtle abnormalities in the sperm, altering their interaction with the Sertoli cells.
FIG. 4.
Retention of mature spermatids in tubules at stages IX to XI in testes of wild-type (circles) and Tnp2 mutant (triangles) mice. The fractions of tubules with 3 to 15 sperm (A), 1 to 2 sperm (B), and no sperm (C) per cross-section are shown. Asterisks indicate values that are significantly different from those for the wild type (P < 0.05). Error bars indicate standard errors.
Epididymal sperm from Tnp2−/− mice showed a significant increase in sperm tail abnormalities over that observed in either wild-type or heterozygote mice (Table 3). Among the tail abnormalities observed in Tnp2−/− mice, the most prominent types were sperm with midpieces that were bent back on themselves (increased from 5% in the wild type to 26%) and sperm in which the axoneme and/or outer dense fibers were unraveled (increased from 2 to 14%). In contrast, there was no increase in the percentage of sperm with head abnormalities, including heads with blunt tips, which were previously found in Tnp1−/− mice (62).
Altered protein levels in Tnp2 mutant mice.
TP1 and TP2 levels in the 3.5% TCA-soluble protein extracts of total testis were analyzed after separation by gel electrophoresis. Although quantification of Coomassie blue-stained bands was valid for measuring the levels of TP1 (62), there were some proteins in Tnp2-null mice, with a total intensity of 23% of that of TP2 from wild-type mice, that migrated in the TP2 region of the gel (Fig. 3A). However, immunoblotting confirmed the absence of TP2 in these Tnp2−/− mice (Fig. 3B). In heterozygotes, the level of TP2 was about 50% of that in wild-type mice. The level of TP1 was elevated by about 45% in Tnp2-null mice as shown by both the Coomassie blue-stained gels and immunoblots (Table 4). TP1 was also elevated in Tnp2 heterozygotes, but to an intermediate level.
TABLE 4.
TP levels in Tnp2 mutant mice
| Tnp2 genotype | Ratio of protein level to that in the wild typea determined by:
|
||
|---|---|---|---|
| Coomassie blue staining (n = 6) (TP1) | Immunoblotting (n = 3)
|
||
| TP1 | TP2 | ||
| Tnp2+/− | 1.22 ± 0.06* | 1.10 ± 0.05 | 0.56 ± 0.06 |
| Tnp2−/− | 1.45 ± 0.10** | 1.40 ± 0.25 | 0 |
All values were first normalized to levels of H1 in the same lane. * and **, significantly different from 1 at P values of <0.05 and <0.01, respectively.
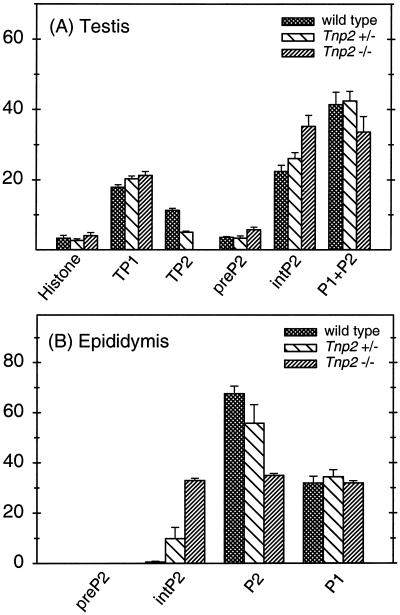
The basic nucleoprotein analysis of step 12 to 16 SRS showed that the level of histones, some of which could result from contamination from other cells, was less than 5% in these cells in both heterozygote and Tnp2-null mice and was not much different from that detected in wild-type mice (Fig 5A and 6A). Furthermore, no histones were detected in extracts from epididymal sperm (Fig. 5B). These results suggest that histones still could be replaced normally without the involvement of TP2.
FIG. 5.
Basic nuclear proteins from sonication-resistant (step 12 to 16) spermatids (A) and epididymal sperm (B) from wild-type, Tnp2+/−, and Tnp2−/− mice. A total basic nuclear protein extract (BNP) from unfractionated wild-type mouse testis nuclei was used as a marker for the histone bands and TP1. The SRS marker represents sonication-resistant step 12 to 16 spermatids prepared from the testes of wild-type mice. In panel B, the heterozygote samples were overloaded on this gel so that the low level of P2 precursors would be clearly visible. Parts of the epididymal preparations were aminoethylated to separate P1 and the mature form of P2. pre-P2 is the primary translation product of the Prm2 gene; pre-P2/11, pre-P2/16, pre-P2/20, and pre-P2/32 result from proteolytic cleavage before the 11th, 16th, 20th, and 32nd amino acids, respectively.
FIG. 6.
Basic nuclear proteins in sonication-resistant spermatids (A) and epididymal sperm (B) as a percentage of total basic protein. Error bars indicate range (n = 2).
In wild-type mice, 11% of the total basic nuclear proteins from SRS was TP2, whereas in heterozygotes it was only 5% and none was detected in the Tnp2-null mice (Fig. 5 and 6). Within the population of step 12 to 16 spermatids, the step 12 to 13 spermatids are the ones that normally contain TP2; hence, these changes most likely reflect events in these cells.
In contrast, the step 14 to 16 spermatids normally contain almost exclusively the protamines. In wild-type or heterozygous mice, 5% of the protamines from SRS were in the form of the P2 precursor pre-P2, about 35% were in the partially processed forms int-P2, and about 60% were in the mature forms P1 and P2 (calculated based on the data in Fig. 6). However, in Tnp2-null mice, the proportion in the mature forms was reduced to 45%, and there was an increase in the partially processed forms, especially pre-P2/11 (Fig. 5A).
Although in epididymal sperm from wild-type mice P2 processing was essentially complete, with 99% of P2 in mature form, in Tnp2-null mice partially processed P2 was still present, with the predominant forms being pre-P2/16 and pre-P2/20 (Fig. 5B). Still, the fraction of P2 in the intermediate forms was 49% of the total amount of P2. Processing was also incomplete in the Tnp2 heterozygotes, with 15% of P2 in intermediate forms.
Even though the processing of P2 was affected by the Tnp2-null mutation, the proportion of total protein that was P2 was not. This is reflected by the percentage of total protamine that was P1, which remained at about 32% in mice with Tnp2 mutations (Fig. 6B).
Altered chromatin condensation in Tnp2 mutant spermatids and sperm.
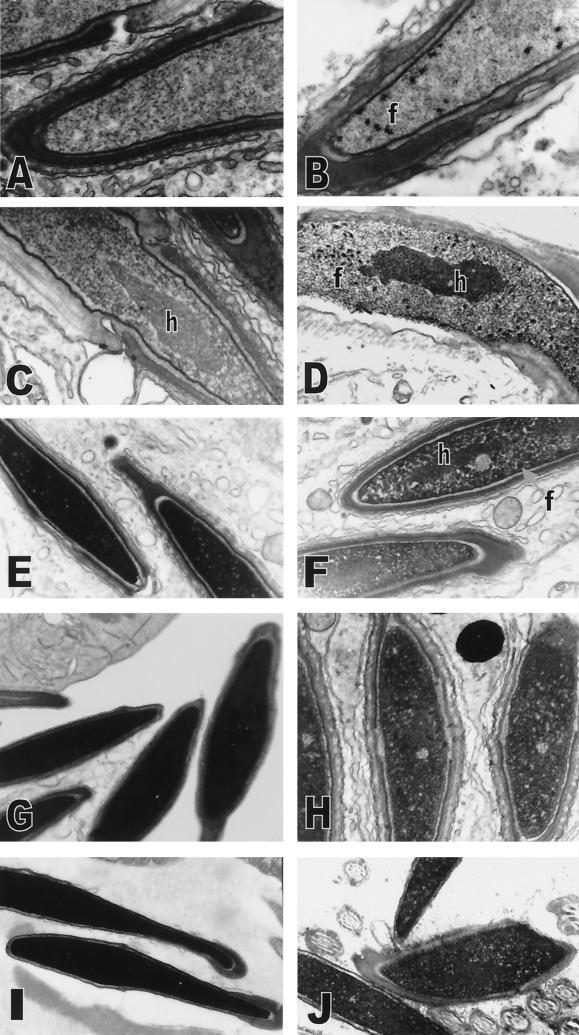
The first abnormalities in chromatin condensation in Tnp2−/− mice were focal condensations, which appeared cylindrical, averaging 95 nm long and 65 nm across as measured in thin sections of randomly oriented units. They were in all late step 11 spermatids (Fig 7B), a stage in which there was no condensation in wild-type spermatids (Fig. 7A). These abnormal condensations appeared primarily in the anterior part of the nucleus. The remainder of the chromatin appeared as a fine needle-like network both in wild-type and Tnp2−/− mice. During steps 12 to 13, these focal condensations increased in number and, in some cases, were found throughout the entire spermatid nucleus (Fig. 7D). The differentially condensed core of chromatin representing the pericentromeric heterochromatin (25), which developed and increased in density during steps 11 to 12 in wild-type mice (Fig. 7C), was similarly observed in spermatids from Tnp2−/− mice (Fig. 7D). During steps 12 to 13 in both wild-type and Tnp2−/− mice, the chromatin fibrils similarly thickened and appeared more rope-like, and the fibrils also moved closer to each other in the process of condensing the chromatin and decreasing the nuclear volume (Fig. 7C and D), sometimes with a gradient from the anterior to the posterior end (not shown). In wild-type mice, the most dramatic condensation occurred as the spermatids progressed through step 13 and into step 14 (Fig. 7E). However, in step 14 spermatids from Tnp2−/− mice, the chromatin, consisting of fibrils and focal condensations, had not completely compacted (Fig. 7F), and occasionally the focal condensations and the differentially condensed pericentromeric heterochromatin remained distinguishable (Fig. 7F). In wild-type mice, the chromatin became uniformly dense when spermatids reached step 15 (not shown), and its appearance by electron microscopy did not change further during step 16 (Fig. 7G) or epididymal maturation (Fig. 7I). However, step 16 spermatids from Tnp2−/− mice still displayed incomplete condensation with lacunae, i.e., small spaces scattered between the dense chromatin, and occasionally a larger area that lacked condensed material (Fig. 7H). These areas of incompletely condensed chromatin were further reduced, but still present, in sperm from the cauda epididymis and vas deferens (Fig. 7J). The chromatin appeared to be arranged in randomly but closely packed spherical objects, with an 11-nm average diameter and an 18-nm average spacing, evenly distributed throughout the nucleus.
FIG. 7.
Abnormalities in chromatin condensation in spermatids from Tnp2 mutant mice. (A and B) Step 11 spermatids from wild-type (A) and Tnp2−/− (B) mice. (C and D) Step 13 spermatids from wild-type (C) and Tnp2−/− (D) mice. (E and F) Step 14 spermatids in stage II from wild-type mice (E) and in stage III from Tnp2−/− mice (F). (G and H) Step 16 spermatids in stage VIII from wild-type mice (G) and in stage VII from Tnp2−/− mice (H). (I and J) Cauda epididymal sperm nuclei from wild-type mice (I) and sperm nuclei from the vas deferens from Tnp2−/− mice (J). f, abnormal focal condensations; h, differentially condensed core of pericentromeric heterochromatin. Magnifications: A, ×25,000; B, ×16,000; C, ×31,000; D, ×10,000; E, ×12,000; F, ×13,000; G, ×11,000; H, ×16,000; I, ×12,000; J, ×12,000.
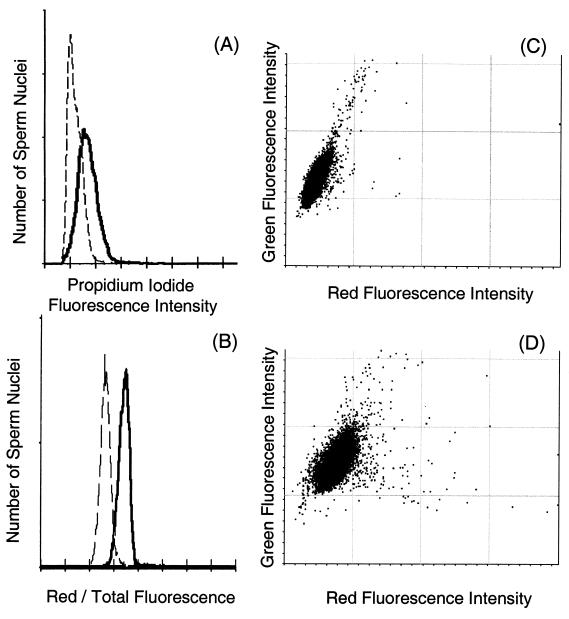
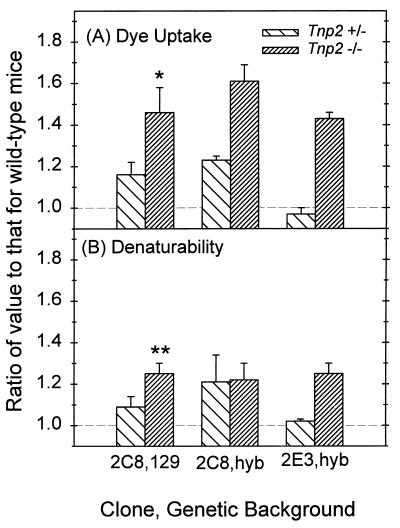
The uptake of intercalating dyes, as measured by the fluorescence from propidium iodide, was also used to assess chromatin condensation (Fig. 8). Tnp2-null mice showed a significant 46% increase in fluorescence over that observed with wild-type sperm nuclei (Fig. 8A and 9A). Similar results were obtained when the absolute amount of green fluorescence with AO was measured, a value that also represents the ability of the dye to intercalate in DNA (Fig 8C and D). Qualitatively, the same effects were seen in trypsin- and sonication-prepared nuclei, although the trypsinized samples showed a greater proportional increase in dye uptake when the Tnp2 genes were eliminated.
FIG. 8.
Flow cytometric analysis of sperm chromatin defects. (A) Distributions of propidium iodide fluorescence intensities for sperm heads prepared by sonication from Tnp2-null mice (solid line) and from wild-type mice (dashed line). (B) Distributions of the relative amount of red fluorescence (red/total fluorescence) of acid-denatured, AO-stained sperm heads prepared with trypsin from Tnp2-null mice (solid line) and from wild-type mice (dashed line). (C and D) Bivariate histograms of green versus red fluorescence for acid-denatured, AO-stained sperm heads prepared by sonication from wild-type (C) and Tnp2-null (D) mice.
FIG. 9.
Dye uptake and susceptibility to acid denaturation of DNA in cauda epididymal sperm nuclei from Tnp2 mutant mice on different genetic backgrounds and derived from different ES cell clones. All values were normalized to those for wild-type mice derived from the same matings as were the mutants. (A) Propidium iodide fluorescence intensity. (B) Denaturability as defined by the ratio of red fluorescence to total (red plus green) fluorescence of AO-stained sperm nuclei. For clone 2C8 on the 129 background, n = 3 to 5; for clone 2C8 and clone 2E3 on the hybrid (hyb) background, n = 2. When n = 2, the error bars represent the range of values; otherwise the error bars indicate standard errors. ∗ and ∗∗, significantly different from 1 at P values of <0.05 and <0.005, respectively.
Susceptibility of sperm DNA from Tnp2 mutants to acid denaturation.
The metachromatic fluorescence of sperm nuclei measured after AO staining and acid denaturation showed that elimination of the Tnp2 genes resulted in an increase of green fluorescence of only 16% but an increased of red fluorescence of 62% (Fig. 8C and D). The ratio of red to total fluorescence intensity was increased significantly in sperm from Tnp2-null mice over that observed in wild-type mice (Fig. 8B and 9B), indicating a greater susceptibility of the DNA to denaturation. The increase was homogenous, as there was no significant broadening of the distribution as measured by its coefficient of variation.
No qualitative differences with different genetic backgrounds, clones, and removal of neo.
Since the effects of the null mutation for Tnp2 could be different on various genetic backgrounds, we examined whether the abnormal phenotypes observed in 129 mice were also present in F2 hybrid (129/B6) mice. Tnp2-null hybrid mice showed the same trends towards reduced litter size and abnormal sperm tail morphology as did the ones on the 129 background (Table 3), but on the 129/B6 hybrid background, the differences from wild type were not statistically significant. In sperm from the Tnp2-null mice on the hybrid genetic background, the levels of P2 precursors were still elevated, at 17% of the total protamine. This value was above control levels (<1%) although not as high as the 33% value found in sperm from Tnp2−/− mice on the 129 background (Fig. 10).
FIG. 10.
Protamine 2 precursor levels (pre-P2 plus int-P2) expressed as percentages of total protamine in Tnp2 mutant mice produced from different ES cell clones, on different genetic backgrounds, and after the excision of the neo gene from the Tnp2 locus. (A) SRS nuclei (step 12 to 16 spermatids) from testes. (B) Epididymal sperm nuclei. Note that pre-P2 was not present in epididymal sperm of any genotype studied here and that there was <1% int-P2 in epididymal sperm from all wild-type mice. hyb, hybrid. Error bars indicate range (n = 2).
We then examined whether similar results would be obtained from different ES cell clones. Because of a lower frequency of germ line transmission, mice derived from a chimera generated with clone 2E3 were available only on a mixed 129/B6 background. In Tnp2-null mice resulting from clone 2E3, partially processed forms of P2 also persisted in SRS and in epididymal sperm, in which 15% of the total protamine was incompletely processed (Fig. 10).
In order to rule out the possibility that the presence of neo at the targeted Tnp2 locus could affect the transcription of the closely linked protamine genes, testes from mice with the neo gene removed were examined. However, Northern blotting showed that deletion of neo did not have much effect on the transcription of Prm1 and Prm2 (Table 2). Although there appeared to be an elevation of Prm1 mRNA caused by removal of neo in the Tnp2−/− mice when cyclophilin was used as the standard, this increase was not significant when the levels were normalized to Tnp1 mRNA levels. Another defect, the presence of incompletely processed P2 in epididymal sperm, was still observed in the Tnp2−/− mice after the removal of the neo sequence and accounted for 12% of the total protamine (Fig. 10B).
We also applied sensitive techniques of flow cytometry to evaluate the effects of genetic background and ES cell clone on sperm from Tnp2 mutant mice. When epididymal sperm from Tnp2−/− mice from different genetic backgrounds and ES cell clones were analyzed, the increases above wild-type levels in propidium iodide dye uptake and relative AO red fluorescence were not significantly different from those observed with clone 2C8 on the 129 background (Fig. 9). Sperm from Tnp2+/− mice, at least from the 2C8 clone, showed slight but nonsignificant increases in dye uptake and denaturability compared to the wild type.
Overall, these results showed that although there were some quantitative differences between different genetic backgrounds, ES cell clones, and sequences of the Tnp2-null allele, there were no qualitative differences.
DISCUSSION
To investigate the role of TP2 in mammalian spermiogenesis, we generated a null mutation in the Tnp2 gene using homologous recombination. Mice homozygous for the mutation completely lacked TP2 in their testes. However, the Tnp2 mutation did not severely impair spermiogenesis; many aspects were normal, but others showed minor defects as discussed below. These mice had relatively normal testis histology, normal numbers of mature sperm, and were still fertile. This suggests that TP2 is not essential for spermiogenesis, likely because the negative consequences caused by the absence of TP2 are minimized by compensation initially by TP1 and later by the protamines.
We did not detect any defects in histone displacement in Tnp2 mutant mice. This result supports our suggestion, based on similar observations with Tnp1-null mice, that histone displacement may be an active process and may start before the initiation of TP accumulation in the nucleus (62).
The transcription of the genes for the other basic nuclear proteins (Tnp1, Prm1, and Prm2) was not affected by the Tnp2 mutation, and the levels of Tnp2 mRNA in Tnp2+/− mice were reduced to about half of that found in the wild type. These results indicate that there is no feedback at the mRNA level. Furthermore, these mRNAs are among the last to be transcribed in step 7 to 9 spermatids (40) before the global repression of transcription at step 10 (30). Therefore, the absence of increased levels of these mRNAs in Tnp2-null mice suggests that TP2 is not responsible for the global repression of transcription.
In contrast to the lack of effect on mRNA levels, TP1 protein levels were elevated by 22 and 45% in Tnp2+/− and Tnp2-null mice, respectively, relative to the wild type. This regulation of TP1 must therefore occur at the posttranscriptional level. These results could be explained by the Tnp1 mRNA being capable of acting as a template for more protein than is produced in wild-type mice and translating more protein because of the reduced amount of TP2. An alternative explanation for the elevated level of TP1 could be its prolonged retention in spermatids beyond step 14. Precedence for this comes from observations of prolonged retention of TP2 in Tarbp2-null mice, which fail to translationally activate the protamines in some cells (63), and in Camk4-null mice, in which P2 is lost from the nucleus soon after deposition (60). However, since protamine levels are not reduced in the Tnp2-null mice, this alternative would appear not to apply.
The Tnp2 mutation also resulted in a deficiency in P2 processing by a mechanism that is not yet clear. Failure of P2 processing has also been observed in Tnp1−/− mice (62), mice that prematurely express P1 at step 8 (37), and mice with haploinsufficiency in either protamine gene (13), suggesting that the normal arrangement of both TPs and protamines on the chromatin is a prerequisite for complete P2 processing. The failure of TP2 removal in spermatids with failure of P2 synthesis (63) or P2 retention and maturation (60) indicates that there may be some interaction between TP2 and P2 by which abnormalities in TP2 levels can affect P2 processing.
Although some aspects of chromatin condensation, such as the thickening of the chromatin fibrils and their condensation during steps 12 and 13, were normal in spermatids from Tnp2-null mice, there were significant abnormalities both in the initiation of condensation and in the final outcome. First was the abnormal focal condensations in step 11 spermatids of Tnp2−/− mice, which were also found in spermatids from Tnp1-null mice, in which they were originally called rod-shaped units and were even more evident (62). The cause of this abnormality might be a consequence of the abnormal deposition of other nuclear proteins present in excess levels and/or an unstable chromatin structure caused by lack of TP2.
Later there was a failure of completion of chromatin compaction in Tnp2 mutant sperm. This was shown by the lacunae in step 15 to 16 spermatids, the granular appearance of the chromatin in epididymal sperm, and the increased uptake of intercalating dyes by the sperm nuclei. The level of intercalating dye fluorescence appears to be primarily related not to the protein composition of the nucleus (17) but rather to the degree of disulfide cross-linking of the protamine (10, 19, 48). Although propidium iodide dye uptake appears to measure a different aspect of chromatin condensation than does electron microscopy, the changes observed when Tnp2 is mutated vary in parallel. Thus, although TP2 is not essential for chromatin condensation, there are defects in the ultimate degree of condensation in Tnp2-null mice.
In addition to the incomplete condensation of the chromatin, there was an increase in DNA denaturation as indicated by the higher relative red fluorescence of AO-stained sperm from Tnp2-null mice. An increase in the relative red fluorescence was also observed in sperm from mice with haploinsufficiency for either of the protamine genes (13). The increased denaturability of the DNA is believed to result from DNA strand breakage, based on correlations between the numbers of cells with high red fluorescence and other measures of strand breakage (3, 23, 51) and the lack of correlation with protamine disulfide cross-linking (50) or phosphorylation (22).
Despite these defects in chromatin condensation, no significant alterations in sperm nuclear shape were observed in Tnp2−/− mice. It was surprising that the absence of TP2 affected tail structures of epididymal sperm to a greater extent than it did sperm heads, given that TP2 is a nuclear protein. The most common tail defect, a sharp bend in the midpiece, resulting in the flagellum being bent back on itself, was also observed in sperm from Tnp1-null mice and mice with haploinsufficiency for one of the protamines (13). Thus, it appears more likely that the tail defects are due to an indirect secondary effect caused by the absence of TP2 or the presence of incompletely processed P2, rather than a direct cytoplasmic role for TP2.
It is interesting that similar phenotypes were observed in both Tnp2 and Tnp1 mutant mice and in mice with haploinsufficiency for the protamines. Some of the abnormalities, such as the abnormal focal chromatin condensations, deficiency of P2 processing, sperm head abnormalities, and loss of fertility, were less marked in Tnp2-null than in Tnp1-null mice (62). The lower levels of defects observed in the Tnp2-null mice are consistent with TP2 being present in spermatids in smaller amounts than TP1 and being a less conserved protein than TP1 (29, 34). In contrast, the loss of even one of the protamine genes appeared to have greater impact, resulting in a complete failure to produce fertile sperm (13).
The role of TP2 in normal spermiogenesis and the exact mechanism by which a mutation in the Tnp2 gene results in defective spermiogenesis are not known, but the present study provides some information. The similarities of the phenotypes of the Tnp1−/− and Tnp2−/− mutants suggest that cooperative participation of both TP1 and TP2 may be required for completely normal spermiogenesis. Alternatively, the severity of the defect may be primarily dependent just on the dosage of the four basic protein genes, with loss of a protamine gene being most severe and loss of the Tnp2 genes being least severe.
The common mechanism underlying the chromatin abnormalities and sperm tail defects in Tnp1-null, in Tnp2-null, and apparently in protamine-haploinsufficient mice is not known. However, all are deficient in P2 processing, indicating that it may have a central role in the sperm abnormalities and infertility. This result is consistent with several studies that have implicated reduced levels of mature P2 in sperm, sometimes accompanied by elevation of the levels of P2 precursors, in human infertility (5, 9, 16), but the murine models will allow more rigorous study of a role for maturation of P2 in fertility. Based on sperm abnormalities and reduced sperm motility in Tnp mutant mice, it is probable that the reduced fertility is a result of failure to fertilize the ova, but the possibility remains that there is some postfertilization developmental defect of sperm nuclei containing incompletely processed P2.
The generation of single Tnp1 and Tnp2 mutations has been helpful in understanding the respective roles of TP1 and TP2 in spermiogenesis. However, due to the compensation between proteins, analysis of the single mutations is not sufficient to completely appreciate the roles of the TPs during mammalian spermiogenesis. Therefore, studies of Tnp1 Tnp2 double mutants are needed and are in progress.
ACKNOWLEDGMENTS
We thank Robert Braun for discussions leading to these studies, Kenneth Kleene and Miles Wilkinson for cDNA plasmids, Stephen Kistler and Sylviane Muller for antisera, Kuriakose Abraham for histological preparations, Nguyen T. Van of the University of Texas M.D. Anderson Cancer Center Flow Cytometry Core Facility and Don Evenson and Lorna Jost of South Dakota State University for advice and assistance in establishing the sperm DNA denaturability assay, Marina Konopleva and Nalini Patel for technical assistance with the flow cytometry, and Walter Pagel for editorial assistance.
This work was supported by National Institutes of Health Research grant HD-16843 and core grant CA-16672.
Footnotes
This paper is dedicated to the memory of Lonnie Russell and his many contributions to our understanding of spermatogenesis.
REFERENCES
- 1.Alfonso P J, Kistler W S. Immunohistochemical localization of spermatid nuclear transition protein 2 in the testes of rats and mice. Biol Reprod. 1993;48:522–529. doi: 10.1095/biolreprod48.3.522. [DOI] [PubMed] [Google Scholar]
- 2.Arango N A, Lovell-Badge R, Behringer R R. Targeted mutagenesis of the endogenous mouse Mis gene promoter: in vivo definition of genetic pathways of vertebrate sexual development. Cell. 1999;99:409–419. doi: 10.1016/s0092-8674(00)81527-5. [DOI] [PubMed] [Google Scholar]
- 3.Aravindan G R, Bjordahl J, Jost L K, Evenson D P. Susceptibility of human sperm to in situ DNA denaturation is strongly correlated with DNA strand breaks identified by single-cell electrophoresis. Exp Cell Res. 1997;236:231–237. doi: 10.1006/excr.1997.3719. [DOI] [PubMed] [Google Scholar]
- 4.Balhorn R, Gledhill B L, Wyrobek A J. Mouse sperm in chromatin proteins: quantitative isolation and partial characterization. Biochemistry. 1977;16:4074–4080. doi: 10.1021/bi00637a021. [DOI] [PubMed] [Google Scholar]
- 5.Balhorn R, Reed S, Tanphaichitr N. Aberrant protamine 1/protamine 2 ratios in sperm of infertile human males. Experientia. 1988;44:52–55. doi: 10.1007/BF01960243. [DOI] [PubMed] [Google Scholar]
- 6.Balhorn R, Weston S, Thomas C, Wyrobek A J. DNA packaging in mouse spermatids: synthesis of protamine variants and four transition proteins. Exp Cell Res. 1984;150:298–308. doi: 10.1016/0014-4827(84)90572-x. [DOI] [PubMed] [Google Scholar]
- 7.Baskaran R, Rao M R S. Mammalian spermatid-specific protein TP2 with nucleic acids, in vitro. A comparative study with TP1. J Biol Chem. 1990;265:21039–21047. [PubMed] [Google Scholar]
- 8.Bellvé A R. Purification and characterization of mouse protamines P1 and P2. Amino acid sequence of P2. Biochemistry. 1988;27:2890–2897. doi: 10.1021/bi00408a034. [DOI] [PubMed] [Google Scholar]
- 9.Belokopytova I A, Kostyleva E I, Tomilin A N, Vorob'ev V I. Human male infertility may be due to a decrease of the protamine P2 content in sperm chromatin. Mol Reprod Dev. 1993;34:53–57. doi: 10.1002/mrd.1080340109. [DOI] [PubMed] [Google Scholar]
- 10.Bottiroli G, Croce A C, Pellicciari C, Ramponi R. Propidium iodide and the thiol-specific reagent DACM as a dye pair for fluorescence resonance energy transfer analysis: an application to mouse sperm chromatin. Cytometry. 1994;15:106–116. doi: 10.1002/cyto.990150204. [DOI] [PubMed] [Google Scholar]
- 11.Bucci L R, Brock W A, Meistrich M L. Distribution and synthesis of histone 1 subfractions during spermatogenesis in the rat. Exp Cell Res. 1982;140:111–118. doi: 10.1016/0014-4827(82)90162-8. [DOI] [PubMed] [Google Scholar]
- 12.Caron N, Veilleux S, Boissonneault G. Stimulation of DNA repair by the spermatidal TP1 protein. Mol Reprod Dev. 2001;58:437–443. doi: 10.1002/1098-2795(20010401)58:4<437::AID-MRD12>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 13.Cho C, Willis W D, Goulding E H, Jung-Ha H, Choi Y-C, Hecht N B, Eddy E M. Haploinsufficiency of protamine-1 or -2 causes infertility in mice. Nat Genet. 2001;28:82–86. doi: 10.1038/ng0501-82. [DOI] [PubMed] [Google Scholar]
- 14.Cole K D, Kistler W S. Nuclear transition protein 2 (TP2) of mammalian spermatids has a very basic carboxyl terminal domain. Biochem Biophys Res Commun. 1987;147:437–442. doi: 10.1016/s0006-291x(87)80140-7. [DOI] [PubMed] [Google Scholar]
- 15.Debarle M, Martinage A, Sautiere P, Chevaillier P. Persistence of protamine precursors in mature sperm nuclei of the mouse. Mol Reprod Dev. 1995;40:84–90. doi: 10.1002/mrd.1080400111. [DOI] [PubMed] [Google Scholar]
- 16.de Yebra L, Ballescá J-L, Vanrell J A, Corzett M, Balhorn R, Oliva R. Detection of P2 precursors in the sperm cells of infertile patients who have reduced protamine P2 levels. Fertil Steril. 1998;69:755–759. doi: 10.1016/s0015-0282(98)00012-0. [DOI] [PubMed] [Google Scholar]
- 17.Evenson D, Darzynkiewicz Z, Jost L, Janca F, Ballachey B. Changes in accessibility of DNA to various fluorochromes during spermatogenesis. Cytometry. 1986;7:45–53. doi: 10.1002/cyto.990070107. [DOI] [PubMed] [Google Scholar]
- 18.Evenson D, Jost L. Sperm chromatin structure assay: DNA denaturability. Methods Cell Biol. 1994;42:159–176. doi: 10.1016/s0091-679x(08)61073-0. [DOI] [PubMed] [Google Scholar]
- 19.Evenson D P, Baer R K, Jost L K. Flow cytometric analysis of rodent epididymal spermatozoal chromatin condensation and loss of free sulfhydryl groups. Mol Reprod Dev. 1989;1:283–288. doi: 10.1002/mrd.1080010409. [DOI] [PubMed] [Google Scholar]
- 20.Evenson D P, Jost L K, Baer R K, Turner T W, Schrader S M. Individuality of DNA denaturation patterns in human sperm as measured by the sperm chromatin structure assay. Reprod Toxicol. 1991;5:115–125. doi: 10.1016/0890-6238(91)90039-i. [DOI] [PubMed] [Google Scholar]
- 21.Gendron-Maguire M, Gridley T. Identification of transgenic mice. Methods Enzymol. 1993;225:794–799. doi: 10.1016/0076-6879(93)25051-3. [DOI] [PubMed] [Google Scholar]
- 22.Golan R, Shochat L, Weissenberg R, Soffer Y, Marcus Z, Oschry Y, Lewin L M. Evaluation of chromatin condensation in human spermatozoa: a flow cytometric assay using acridine orange staining. Mol Hum Reprod. 1997;3:47–54. doi: 10.1093/molehr/3.1.47. [DOI] [PubMed] [Google Scholar]
- 23.Gorczyca W, Traganos F, Jesionowska H, Darzynkiewicz Z. Presence of DNA strand breaks and increased sensitivity of DNA in situ to denaturation in abnormal human sperm cells: analogy to apoptosis of somatic cells. Exp Cell Res. 1993;207:202–205. doi: 10.1006/excr.1993.1182. [DOI] [PubMed] [Google Scholar]
- 24.Grimes S R, Jr, Meistrich M L, Platz R D, Hnilica L S. Nuclear protein transitions in rat testis spermatids. Exp Cell Res. 1977;110:31–39. doi: 10.1016/0014-4827(77)90266-x. [DOI] [PubMed] [Google Scholar]
- 25.Haaf T, Ward D C. Higher order nuclear structure in mammalian sperm revealed by in situ hybridization and extended chromatin fibers. Exp Cell Res. 1995;219:604–611. doi: 10.1006/excr.1995.1270. [DOI] [PubMed] [Google Scholar]
- 26.Hecht N B, Kleene K C, Yelick P C, Johnson P A, Pravtcheva D D, Ruddle F H. Mapping of haploid expressed genes: genes for both mouse protamines are located on chromosome 16. Somat Cell Mol Genet. 1986;12:203–208. doi: 10.1007/BF01560667. [DOI] [PubMed] [Google Scholar]
- 27.Heidaran M A, Showman R M, Kistler W S. A cytochemical study of the transcriptional and translational regulation of nuclear transition protein 1 (TP1), a major chromosomal protein of mammalian spermatids. J Cell Biol. 1988;106:1427–1433. doi: 10.1083/jcb.106.5.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horan G S, Kovacs E N, Behringer R R, Featherstone M S. Mutations in paralogous Hox genes result in overlapping homeotic transformations of the axial skeleton: evidence for unique and redundant function. Dev Biol. 1995;169:359–372. doi: 10.1006/dbio.1995.1150. [DOI] [PubMed] [Google Scholar]
- 29.Keime S, Kumm S, Luerssen H, Engel W. The nucleotide sequence of boar transition protein 2 (TNP2) cDNA and haploid expression of the gene during spermatogenesis. Anim Genet. 1992;23:373–378. doi: 10.1111/j.1365-2052.1992.tb00161.x. [DOI] [PubMed] [Google Scholar]
- 30.Kierszenbaum A L, Tres L L. Structural and transcriptional features of the mouse spermatid genome. J Cell Biol. 1975;65:258–270. doi: 10.1083/jcb.65.2.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kistler W S, Henriksén K, Mali P, Parvinen M. Sequential expression of nucleoproteins during rat spermiogenesis. Exp Cell Res. 1996;225:374–381. doi: 10.1006/excr.1996.0188. [DOI] [PubMed] [Google Scholar]
- 32.Kleene K C, Borzorgzadeh A, Flynn J F, Yelick P C, Hecht N B. Nucleotide sequence of a cDNA clone encoding mouse transition protein 1. Biochim Biophys Acta. 1988;950:215–220. doi: 10.1016/0167-4781(88)90013-9. [DOI] [PubMed] [Google Scholar]
- 33.Kleene K C, Flynn J F. Characterization of a cDNA clone encoding a basic protein, TP2, involved in chromatin condensation during spermiogenesis in the mouse. J Biol Chem. 1987;262:17272–17277. [PubMed] [Google Scholar]
- 34.Kremling H, Luerssen H, Adham I M, Klemm U, Tsaousidon S, Engel W. Nucleotide sequences and expression of cDNA clones for boar and bull transition protein 1 and its evolutionary conservation in mammals. Differentiation. 1989;40:184–190. doi: 10.1111/j.1432-0436.1989.tb00597.x. [DOI] [PubMed] [Google Scholar]
- 35.Krishnamurthy H, Danilovich N, Morales C R, Sairam M R. Qualitative and quantitative decline in spermatogenesis of the follicle-stimulating hormone receptor knockout (FORKO) mouse. Biol Reprod. 2000;62:1146–1159. doi: 10.1095/biolreprod62.5.1146. [DOI] [PubMed] [Google Scholar]
- 36.Kundu T K, Rao M R S. Zinc dependent recognition of a human CpG island sequence by the mammalian spermatidal protein TP2. Biochemistry. 1996;35:15626–15632. doi: 10.1021/bi961271i. [DOI] [PubMed] [Google Scholar]
- 37.Lee K, Haugen H S, Clegg C H, Braun R E. Premature translation of protamine 1 mRNA causes precocious nuclear condensation and arrests spermatid differentiation in mice. Proc Natl Acad Sci USA. 1995;92:12451–12455. doi: 10.1073/pnas.92.26.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levesque D, Veilleux S, Caron N, Boissonneault G. Architectural DNA-binding properties of the spermatidal transition proteins 1 and 2. Biochem Biophys Res Commun. 1998;252:602–609. doi: 10.1006/bbrc.1998.9687. [DOI] [PubMed] [Google Scholar]
- 39.Luerssen H, Maier W M, Hoyer-Fender S, Engel W. The nucleotide sequence of rat transition protein 2 (TP2) cDNA. Nucleic Acids Res. 1989;17:3585. doi: 10.1093/nar/17.9.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mali P, Kaipia A, Kangasniemi M, Toppari J, Sandberg M, Hecht N B, Parvinen M. Stage-specific expression of nucleoprotein mRNAs during rat and mouse spermiogenesis. Reprod Fertil Dev. 1989;1:369–382. doi: 10.1071/rd9890369. [DOI] [PubMed] [Google Scholar]
- 41.Meetei A R, Ullas K S, Rao M R S. Identification of two novel zinc finger modules and nuclear localization signal in rat spermatidal protein TP2 by site-directed mutagenesis. J Biol Chem. 2000;275:38500–38507. doi: 10.1074/jbc.M002734200. [DOI] [PubMed] [Google Scholar]
- 42.Meistrich M L. Histone and basic nuclear protein transitions in mammalian spermatogenesis. In: Hnilica L S, Stein G S, Stein J L, editors. Histones and other basic nuclear proteins. Orlando, Fla: CRC Press; 1989. pp. 165–182. [Google Scholar]
- 43.Meistrich M L. Nuclear morphogenesis during spermiogenesis. In: Kretser D M, editor. Molecular biology of the male reproductive system. San Diego, Calif: Academic Press; 1993. pp. 67–97. [Google Scholar]
- 44.Meistrich M L, Trostle-Weige P K, van Beek M E A B. Separation of specific stages of spermatids from vitamin A-synchronized rat testes for assessment of nucleoprotein changes during spermiogenesis. Biol Reprod. 1994;51:334–344. doi: 10.1095/biolreprod51.2.334. [DOI] [PubMed] [Google Scholar]
- 45.Millette C F, Spear P G, Gall W E, Edelman G M. Chemical dissection of mammalian spermatozoa. J Cell Biol. 1973;58:662–675. doi: 10.1083/jcb.58.3.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oko R J, Jando V, Wagner C L, Kistler W S, Hermo L S. Chromatin reorganization in rat spermatids during the disappearance of testis-specific histone, H1t, and the appearance of transition proteins TP1 and TP2. Biol Reprod. 1996;94:1141–1157. doi: 10.1095/biolreprod54.5.1141. [DOI] [PubMed] [Google Scholar]
- 47.Platz R D, Meistrich M L, Grimes S R. Low-molecular weight basic proteins in spermatids. Methods Cell Biol. 1977;16:267–316. doi: 10.1016/s0091-679x(08)60107-7. [DOI] [PubMed] [Google Scholar]
- 48.Rhim J A, Connor W, Dixon G H, Harendza C J, Evenson D P, Palmiter R D, Brinster R L. Expression of an avian protamine in transgenic mice disrupts chromatin structure in spermatozoa. Biol Reprod. 1995;52:20–32. doi: 10.1095/biolreprod52.1.20. [DOI] [PubMed] [Google Scholar]
- 49.Russell L, Burguet S. Ultrastructure of Leydig cells as revealed by secondary tissue treatment with a ferrocyanide-osmium mixture. Tissue Cell. 1977;9:751–766. doi: 10.1016/0040-8166(77)90040-4. [DOI] [PubMed] [Google Scholar]
- 50.Said S, Funahashi H, Niwa K. DNA stability and thiol-disulphide status of rat sperm nuclei during epididymal maturation and penetration of oocytes. Zygote. 1999;7:249–254. doi: 10.1017/s0967199499000635. [DOI] [PubMed] [Google Scholar]
- 51.Sailer B L, Jost L K, Evenson D P. Mammalian sperm DNA susceptibility to in situ denaturation associated with the presence of DNA strand breaks as measured by the terminal deoxynucleotidyl transferase assay. J Androl. 1995;16:80–87. [PubMed] [Google Scholar]
- 52.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 53.Schlüter G, Celik A, Obata R, Schlicker M, Hofferbert S, Schlung A, Adham I M, Engel W. Sequence analysis of the conserved protamine gene cluster shows that it contains a fourth expressed gene. Mol Reprod Dev. 1996;43:1–6. doi: 10.1002/(SICI)1098-2795(199601)43:1<1::AID-MRD1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 54.Schlüter G, Kremling H, Engel W. The gene for human transition protein 2: nucleotide sequence, assignment to the protamine gene cluster, and evidence for its low expression. Genomics. 1992;14:377–383. doi: 10.1016/s0888-7543(05)80229-0. [DOI] [PubMed] [Google Scholar]
- 55.Singh J, Rao M R S. Interaction of rat testis protein TP with nucleic acids in vitro: fluorescence quencing, UV absorption, and thermal denaturation studies. J Biol Chem. 1987;262:734–740. [PubMed] [Google Scholar]
- 56.Singh J, Rao M R S. Interaction of rat testis protein, TP, with nucleosome core particle. Biochem Int. 1988;17:701–710. [PubMed] [Google Scholar]
- 57.Sprando R L. Perfusion of the rat testis through the heart using heparin. In: Russell L D, Ettlin R A, Sinha Hikim A P, Clegg E D, editors. Histological and histopathological evaluation of the testis. Clearwater, Fla: Cache River Press; 1990. pp. 277–280. [Google Scholar]
- 58.Sutton K A, Maiti S, Tribley W A, Lindsey J S, Meistrich M L, Bucana C D, Sanborn B M, Joseph D R, Griswold M D, Cornwall G A, Wilkinson M F. Androgen regulation of the Pem homeodomain gene in mice and rat Sertoli and epididymal cells. J Androl. 1998;19:21–30. [PubMed] [Google Scholar]
- 59.Unni E, Meistrich M L. Purification and characterization of the rat spermatid basic nuclear protein TP4. J Biol Chem. 1992;267:25359–25363. [PubMed] [Google Scholar]
- 60.Wu J Y, Ribar T J, Cummings D E, Burton K A, McKnight G S, Means A R. Spermiogenesis and exchange of basic nuclear proteins are impaired in male germ cells lacking Camk4. Nat Genet. 2000;25:448–452. doi: 10.1038/78153. [DOI] [PubMed] [Google Scholar]
- 61.Yelick P C, Balhorn R, Johnson P A, Corzett M, Mazrimas J A, Kleene K C, Hecht N B. Mouse protamine 2 is synthesized as a precursor whereas mouse protamine 1 is not. Mol Cell Biol. 1987;7:2173–2179. doi: 10.1128/mcb.7.6.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu Y E, Zhang Y, Unni E, Shirley C R, Deng J M, Russell L D, Weil M M, Behringer R R, Meistrich M L. Abnormal spermatogenesis and reduced fertility in transition nuclear protein 1-deficient mice. Proc Natl Acad Sci USA. 2000;97:4683–4688. doi: 10.1073/pnas.97.9.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhong J, Peters A H, Lee K, Braun R E. A double-stranded RNA binding protein required for activation of repressed messages in mammalian germ cells. Nat Genet. 1999;22:171–174. doi: 10.1038/9684. [DOI] [PubMed] [Google Scholar]