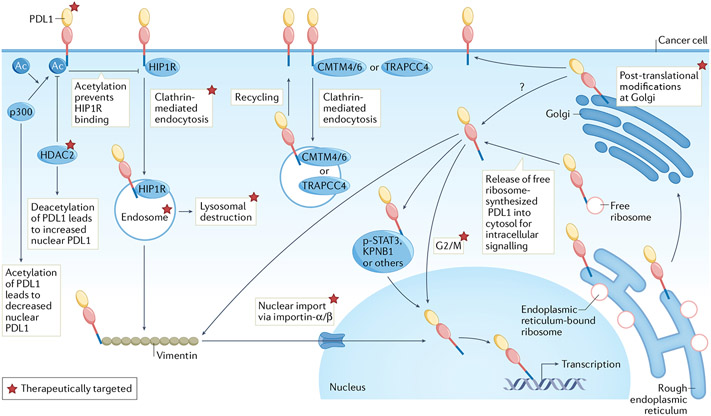
Fig. 2 ∣. Regulation of intracellular PDL1 and potential targeting.
Programmed death ligand 1 (PDL1) is synthesized in endoplasmic reticulum followed by Golgi processing and surface presentation. Huntingtin-interacting protein 1-related protein (HIP1R) targets PDL1 for lysosomal destruction whereas CKLF-like MARVEL transmembrane domain-containing protein 4/6 (CMTM4/6) or trafficking protein particle complex subunit 4 (TRAPPC4) recycles endocytic PDL1 from cytoplasm to surface. HIP1R-dependent clathrin-mediated endocytosis of surface, non-acetylated PDL1 (controlled by histone deacetylase 2 (HDAC2) and acetyltransferase p300) facilitates PDL1 (from free ribosomes) interaction with vimentin followed by nuclear import via importin-α/β. We show p300 and HDAC2 as acting on cell surface PDL1, but those interactions could also occur in cytosol or nucleus. PDL1 can interact with p-STAT3 in triple-negative breast cancer (TNBC) during hypoxia or associate with karyopherin B1 (KPNB1) in lung cancer, either pathway increasing nuclear PDL1. Free ribosomes (that is, not endoplasmic reticulum-bound) can theoretically synthesize PDL1 for direct release into the cytosol without modification in the endoplasmic reticulum and Golgi. Thus, the cellular source of nuclear PDL1 could be from the surface or cytosol, or directly from other organelles following PDL1 processing such as endoplasmic reticulum/Golgi. PDL1 can participate in macromolecular interactions, for example, the endoplasmic reticulum-associated protein degradation pathway, but whether endoplasmic reticulum-bound PDL1 mediates cell-intrinsic signalling is unknown. Apart from acetylation, phosphorylation, glycosylation, palmitoylation and polyubiquitination, other post-translational PDL1 modifications regulate its subcellular distribution, but mechanistic details are not comprehensively characterized. Exogenous ligands and cellular stressors can upregulate PDL1 expression or redistribute surface PDL1 to other locations including the cytosol and nucleus or into organelles such as lysosomes or autophagosomes. Examples include tumour necrosis factor (TNF), interferons, DNA damage (from exogenous sources such as cytotoxic chemotherapy) or environmental cues such as hypoxia or nutrient (for example, glucose) availability. PDL1 cell-intrinsic trafficking is significantly controlled by the cell cycle, decreasing in the G1 phase and increasing in S/G2/M cycles. Regulation of PDL1 cell-intrinsic signalling directly from mitochondria, endoplasmic reticulum, autophagosomes or other non-nuclear organelles remains to be elucidated. Altering cell-intrinsic PDL1 content or signalling apart from surface PD1–PDL1 signal disruption using US Food and Drug Administration (FDA)-approved antibodies elicits distinct therapeutic vulnerabilities. Total tumour PDL1 can be reduced using approaches including agents targeting PDL1 post-translational modifications such as palmitoylation and glycosylation (for example, peptide mimics, glutathione synthase kinase 3β (GSK3β) inhibitors), antibodies disrupting PDL1–CMTM4 or CMTM6 interactions (for example, H1A), repurposed FDA-approved drugs (for example, the photosensitizer verteporfin) and natural products (for example, curcumin). Subcellular PDL1 redistribution can be altered using agents disrupting clathrin-mediated endocytosis (for example, pitstop 2), macromolecular interactions (for example, cytoskeleton or signal transducer and activator of transcription 3 (STAT3) inhibitors) and importin-α/β molecules for nuclear import (for example, FDA-approved ivermectin). Therapies sequestering PDL1 on cell surface or in other compartments such as endosomes or autophagosomes include FDA-approved cell-cycle inhibitors (for example, palbociclib, ribociclib), endocytic inhibitors (for example, Dyngo-4a) and autophagy inhibitors (for example, hydroxychloroquine).