Abstract
Purpose:
To investigate the diagnostic performance of a deep convolutional neural network for differentiation of clear cell renal cell carcinoma (ccRCC) from renal oncocytoma.
Methods:
In this retrospective study, 74 patients (49 male, mean age 59.3) with 243 renal masses (203 ccRCC and 40 oncocytoma) that had undergone MR imaging 6 months prior to pathologic confirmation of the lesions were included. Segmentation using seed placement and bounding box selection was used to extract the lesion patches from T2-WI, and T1-WI pre-contrast, post-contrast arterial and venous phases. Then, a deep convolutional neural network (AlexNet) was fine-tuned to distinguish the ccRCC from oncocytoma. Five-fold cross validation was used to evaluate the Al algorithm performance. A subset of 80 lesions (40 ccRCC, 40 oncocytoma) were randomly selected to be classified by two radiologists and their performance was compared to the AI algorithm. Intra-class correlation coefficient was calculated using the Shrout-Fleiss method.
Results:
Overall accuracy of the AI system was 91% for differentiation of ccRCC from oncocytoma with an area under the curve of 0.9. For the observer study on 80 randomly selected lesions, there was moderate agreement between the two radiologists and AI algorithm. In the comparison sub-dataset, classification accuracies were 81%, 78%, and 70% for AI, radiologist 1, and radiologist 2, respectively.
Conclusion:
The developed AI system in this study showed high diagnostic performance in differentiation of ccRCC versus oncocytoma on multi-phasic MRIs.
Keywords: Deep learning, Radiomics, Clear cell renal cell carcinoma, Oncocytoma, Multi-phasic MRI
1. Introduction
Increased use of cross sectional imaging has resulted in remarkable rise in detection of small renal masses, which are defined as masses with the largest dimension of ≤4 cm on abdominal imaging.1 3 Small renal masses (SRMs) can demonstrate a wide spectrum of prognosis from benign neoplasms and cysts to aggressive malignancies. 4,5
The majority of contrast-enhancing SRMs are Renal Cell Carcinomas (RCC) that encompass a heterogeneous group of cancers, with clear cell subtype (ccRCC) being the most common subclassification of RCCs that accounts for the majority of kidney cancerrelated mortalities.5‘6 While enhancement increases the chance of malignancy, a considerable portion of contrast-enhancing SRMs are benign tumors.7‘8 For instance, Oncocytoma, the most common benign renal mass often demonstrates enhancement patterns similar to ccRCC, making it challenging to differentiate these two subtypes that have different prognostic outcomes.9,10
Due to uncertainty in pre-operative diagnosis and lower incidence, benign tumors tend to be over treated, while active surveillance is an option for many patients with these tumors. In fact, renal oncocytoma accounts for 4–10% of nephrectomies in which renal-cell carcinoma is suspected.11,12
Percutaneous renal mass biopsy is suggested in cases that knowledge of histology could influence subsequent management.13 However, renal mass biopsy is an invasive procedure, associated with complications related to the procedure itself or difficulties with acquiring the representative tissue.14 In addition, the collected biopsy samples may be inadequate or indeterminate in a number of cases. For instance, benign oncocytoma coexists with RCC in 20% of cases of oncocytoma which might lead to imprecise biopsy and false negative results and subsequent detrimental clinical outcomes.14,15
It is evident that in such heterogeneous clinical setting, development of diagnostic tools could have significant impact on patient management and care. However, efforts for finding a robust radiologic method to accurately differentiate ccRCC and oncocytoma, has not yet delivered any finding of high clinical value.16,17
To determine the biologic nature of an observed lesion from imaging data, a common practice for the radiologist includes evaluation of certain morphologic features such as boundary, sphericity, internal structure, and texture. However, these features, which can be quite subjective, suffer from high inter/intra-observer variability. In some more challenging scenarios, such as the present task of classification of oncocytoma and ccRCC, it may not be feasible to design such a rule-based system due to the subtle and hard-to-translate difference between the two. In the past decades, radiomics has enabled us to analyze medical images beyond what human eye is capable of detecting. Radiomics has facilitated automatic detection and characterization of renal masses. Early attempts mostly relied on a combination of handcrafted features and machine learning classifiers to estimate the desired output in tasks such as computer aided diagnosis (CAD). The features were mostly general-purpose initially designed for natural images and were fed to common classifiers such as support vector machine (SVM) and random forest. These methods often suffered from limited accuracy and robustness.18 With the recent development of deep learning and artificial intelligence (AI), robust hierarchical features can be learnt directly from data and tasks which can achieve performance matching to that of human.19,20 This can be especially useful for tasks where the features are too subtle or difficult to catch explicitly.
The purpose of this article was to develop a deep-learning based AI system for differentiation of clear cell renal carcinoma from benign oncocytoma and to evaluate its performance. We further compared the result from the AI algorithm with the assessment of lesion types from two independent radiologists.
2. Methods
2.1. Patient population
This study was compliant with the Health Insurance Portability and Accountability Act (HIPAA) and was approved by our institutions Institutional Review Board (IRB). Patients were recruited consecutively for a prospective study of natural history of hereditary renal cancers at the Urologic Oncology Branch of the National Cancer Institute (UOB-NCI). All patients signed written informed consent to participate in this protocol (NCI-89-C-0086).
Our patient cohort was selected through the retrospective review of a prospectively maintained UOB-imaging registry for individuals with pathological diagnosis of oncocytoma or ccRCC that had undergone surgical resection of tumors between February 2014 and July 2018 and had contrast-enhanced Magnetic Resonance Imaging (MRI) within 6 months prior to pathologic confirmation. Images were acquired on 1.5 T MRI scanners (Aera, Siemens Healthcare) with the same dedicated renal mass protocol. Patients with images that had significant artifact and lesions smaller than 1 cm were excluded from further analysis. Fig. 1 shows the accrual flowchart of our study based on inclusion and exclusion criteria used in this study.
Fig. 1.
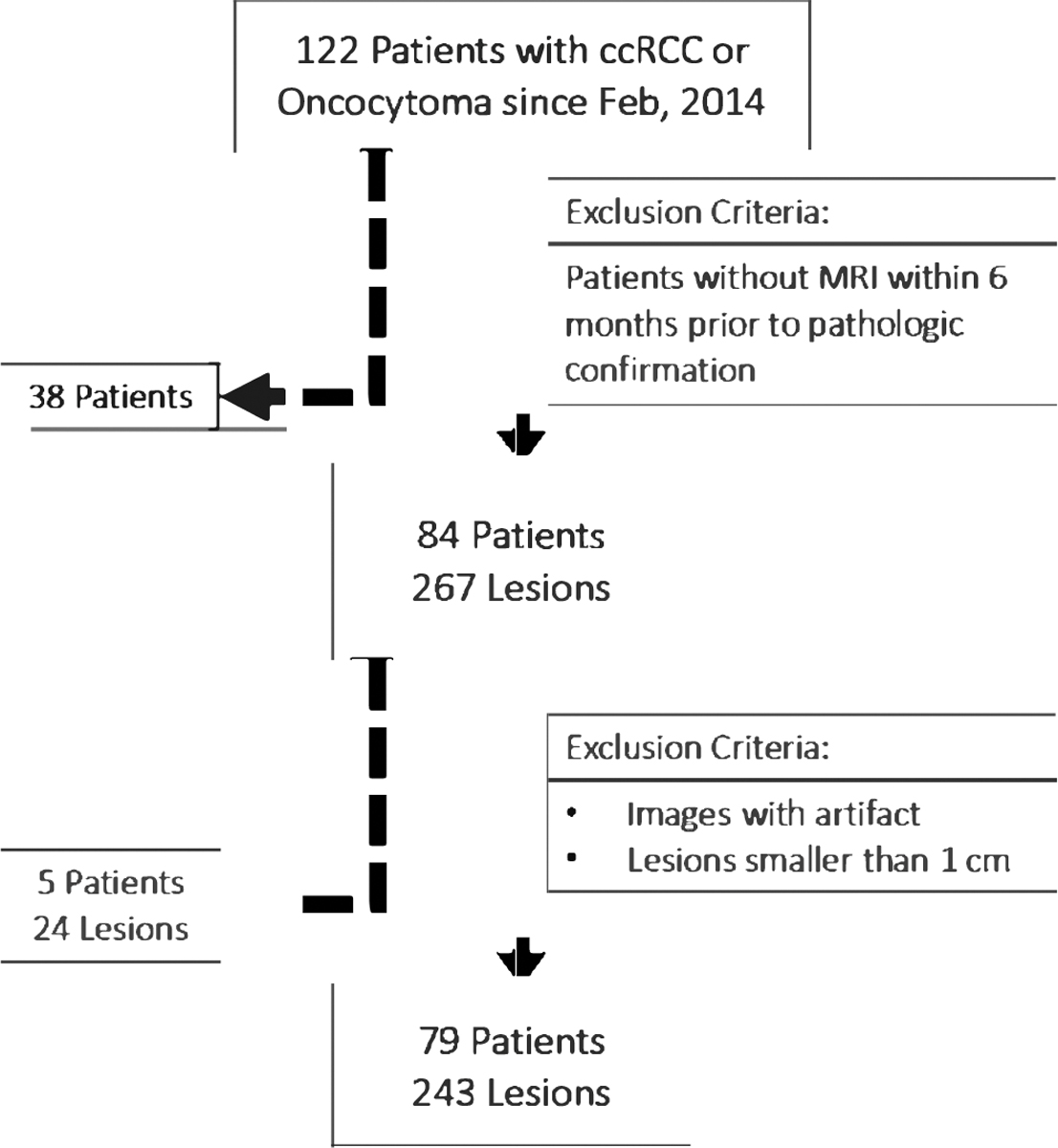
Study flow chart summarizing the patient and lesion selection.
2.2. Magnetic resonance imaging acquisition
MRI examinations were performed per our institution’s protocol for renal masses at a field strength of 1.5 T (Aera, Siemens Healthcare). Images were acquired before and after an administration of gadobutrol (Gadavist, Bayer, Whippany, New Jersey, USA) at 0.1 mmol/kg and 20 mL saline f ush. Imaging sequences included T2-weighted images (T2-WI), and pre- and post contrast T1-weighted Images (T1-WI) during corticomedullary phase (20 s), and nephrographic phase (70 s) (Table 1).
Table 1.
Sequence acquisition parameters for Echo Planar Imaging of renal masses.
| Parameter | T2 WI without fat saturation | Pre-contrast T1 WI | Arterial Phase T1-WI | Venous Phase T1-WI |
|---|---|---|---|---|
|
| ||||
| TR (ms) | 1700 | 3.78 | 3.78 | 3.78 |
| TE (ms) | 112 | 1.74 | 1.74 | 1.74 |
| No. of slices | 40 | 104 | 104 | 104 |
| Slice thickness (mm) | 6 | 3 | 3 | 3 |
| Flip angle (°) | 140 | 10 | 10 | 10 |
| Matrix | 232 × 256 | 270 × 320 | 270 × 320 | 270 × 320 |
| FOV (cm) | 148.5 × 66.9 | 119 × 53.5 | 119 × 53.5 | 119 × 53.5 |
| Voxel size (mm) | 7.8 | 3 | 3 | 3 |
TR: repetition time; TE: echo time; FOV: field of view.
2.3. Image registration
In order to fully exploit the information from different MRI sequences in the AI algorithm, we aligned all the pre-contrast and contrast-enhanced (arterial and venous) sequences of T1-weighted images (T1-WI) with the T2-weighted images (T2-WI). Since patients have limited body or abdomen movement during the MRI scanning, a rigid image registration was sufficient to align all the contrast sequences to the T2-WI. More specifically, we designated the T2 volumetric image as the target image, denoted as , and contrast enhanced volumetric images as the moving images, denoted as Icontrast. The goal was to find the optimal 3D spatial transformation T(.), which maps any point in the moving image Icontrast to its corresponding point in the target image . Mutual information (MI) was used as their similarity measurement, which was defined by , where H(Icontrast) and denoted the marginal entropies of Icontrast and , and denoted their joint entropy that was calculated from their joint histogram. We used the Insight Segmentation and Registration Toolkit (ITK) (https://itk.org/) to perform the image registrations.
2.4. Image processing for training data generation
Oncocytoma and ccRCCs were cropped out as two-dimensional (2D) patches to fine-tune the AI algorithm for their classifications. First, for each lesion, an imaging research fellow with 3 years of experience manually selected a seed point roughly at the lesion center from each lesion slice in T2-WI, using image analysis software ITK-SNAP (v3.6.0).21 Then, local region of interest (ROI) patch was automatically extracted around the seed point with a fixed size of 100 × 100 mm in physical space. After that, corresponding patches from sets of three image series were selected, creating four possible sequence combinations. Image patches from each set were combined as a red green blue (RGB) image and resized to 224 × 224 to fit the pre-trained AlexNet configuration, which requires 3 channels for the input image.
Data Augmentation was performed on training data to increase the size of training sets, and to reduce the overfitting. To balance the training number of ccRCC and oncocytoma lesions (originally approximately 5:1), different ratios of data augmentations were performed. For each oncocytoma lesion patch, 20 rotations with random degree from −90 to 90 were applied. And for each ccRCC lesion patch, 4 rotations with random degree from −90 to 90 were applied. For both types of lesions, a flipping operation from top to down was also applied. Fig. 2 shows an example of image segmentation performed for training data generation.
Fig. 2.

Showing segmentation process and bounding box generated of the tumor shown in Fig. 2. Tumor was located on T2 WI (A), a seed point was placed on the lesion based on rough estimation of its location (B). Bounding box was generated, relative to the image (C), and extracted (D).
2.5. Architecture
Considering the fact that our kidney lesion dataset was relatively small (compared to millions of pictures in natural image domain) and the pre-trained AI algorithms were Jess prone to overfitting, we chose to fine-tune a relative shallower convolutional neural network (CNN), i.e. AlexNet,22 which contains five convolutional layers followed by three fully-connected layers. The AlexNet architecture is presented in Fig. 3. We modified the last fully connected layer from 1000 original neural nodes to 2 nodes representing oncocytoma and ccRCC classes, while keeping other layers setup unchanged. The output of the last filly connected layer is further processed by a two classes softmax function before calculating network losses. The cross-entropy loss was used to train the network.
Fig. 3.
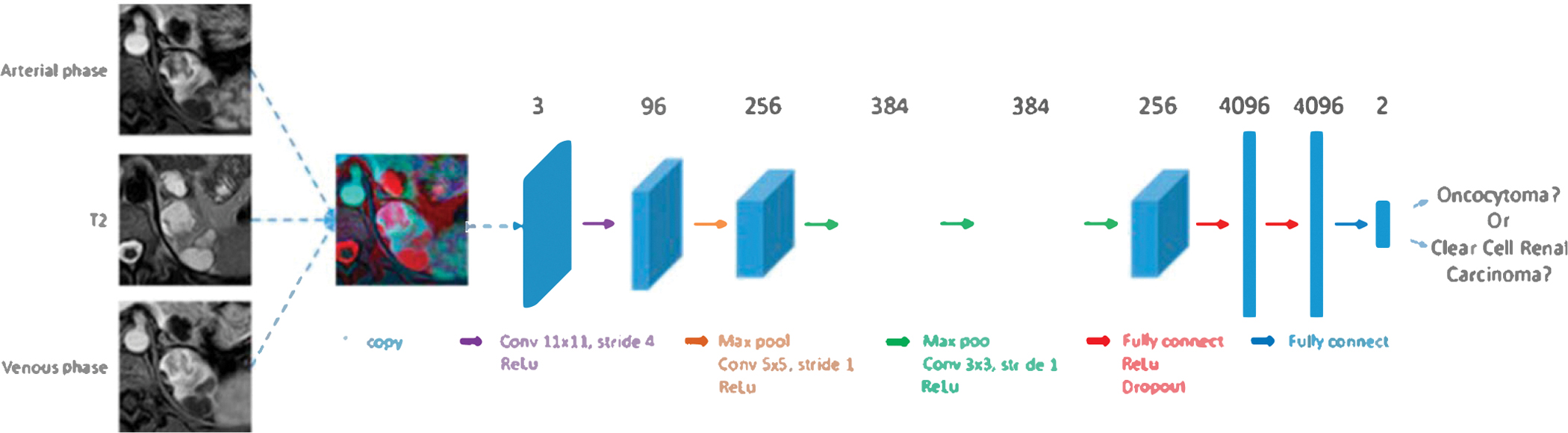
Convolutional neural network architecture.
2.6. Training
We used 5-fold cross validations at lesion level to get a comprehensive evaluation of the classifier’s performance on differentiating oncocytoma and ccRCC lesions. For each fold, there were roughly 160 ccRCC and 32 oncocytoma lesions used for training and 40 ccRCC and 8 oncocytoma lesions for testing. Each lesion contained 1– 10 (average 4.5) slices. After data augmentation, there was a total –7000 training patches for each fold.
We used the deep learning library of Caffe23 to fine-tune the AlexNet, where optimization was performed using stochastic gradient descent, with an initial learning rate of le-6, a momentum of 0.9, and a weight decay of 0.005. The learning rate was reduced by 0.1 after every 40 epochs. Training converged after –100 epochs taking about 2 h in an Nvidia K40 GPU.
2.7. Performance evaluation metrics
The fine-tuned network was used to predict the label of testing data. Final per lesion prediction was obtained by aggregating the probability scores across all lesion slices. Final performance was reported by averaging the prediction results from 5 folds. Receiver operating characteristic (ROC) analysis was performed and area under the curve (AUC) was calculated. Sensitivity, specificity, precision or positive predictive value (PPV) and negative predictive value (NPV), and accuracy in differentiating oncocytoma and ccRCC were calculated.
2.8. Clinical validation
To compare radiologists’ vs. AI performance on this classification task, we designed an observer study that allowed readers to simultaneously view all sequences of pre-contrast and contrast-enhanced (arterial and venous) T1-WI and T2 WI, while being able to scroll through the volume that only included lesion and perilesional information. Images were cropped to the size of the bounding box used for the deep neural network to include only the ipsilateral kidney in order to eliminate confusion of contralateral kidney and were randomly presented to radiologists. Two fellowship-trained radiologists (with 30 and 10 years of experience), evaluated a total of 80 (40 ccRCC, 40 oncocytoma) masses that were randomly selected from the same cohort. Radiologists were blinded to all patient clinical and histology information and were asked to provide their diagnosis on a grade of 1–4 as their certainty of decision. A score of 1 was defined as when the radiologist observed enough evidence (including but not limited to homogeneous, well-circumscribed solid mass, often containing a central scar) to have high confidence about the diagnosis of oncocytoma. A score of 2 was assigned when the radiologist preferred the oncocytoma lesion type but with low confidence to confirm its diagnosis. Similar to the score of 1 and 2 used for oncocytoma lesions, score of 3 was assigned if the radiologist preferred the ccRCC lesion type but with low confidence to confirm its diagnosis, while score of 4 was assigned when the radiologist was highly confident that the lesion type was ccRCC. This scoring system could better reflect the decision process of radiologists and also more comparable to the probability-based outputs from AI algorithm. This scoring system could also allow for a ROC analysis by setting the positive threshold at each individual score, resulting in 5 points within ROC plane.
2.9. Performance, comparison
The intra-class correlation coefficient of the two radiologists and AI classifications was determined by using Shrout Fleiss method. Accuracy, sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) were calculated for the radiologists and were compared to the performance of AI on the comparison subset (80 tumors with 40 oncocytoma and 40 ccRCC lesions). Next, the probability outputs of AI algorithm for these 80 lesions were rounded to the nearest quartile to more precisely investigate probability outcomes rather than a binary decision and to compare to radiologist rating of 1–4. More specifically, if AI’s output probability for oncocytoma was greater than 0.75, the score 1 was assigned meaning the AI algorithm had high confidence that it was an oncocytoma lesion, and if the output probability for oncocytoma was between (0.5, 0.75), the score of 2 was assigned meaning the network had low confidence on suggesting an oncocytoma lesion. Similarly, the score of 3 and 4 were defined if the network had low and high confidence, respectively to predict the lesion as ccRCC type.
3. Results
Our study included a final cohort of 74 patients (49 male, mean age 59.3 years) with 243 renal masses, consisting of 203 ccRCC from 59 patients and 40 oncocytoma from 15 patients. From the four sequence combinations, using T2 WI and T1-post contrast sequences yielded the highest performance as reported in the following.
In the 5-fold cross validation, AI showed sensitivity of 94%, specificity of 75% and an overall accuracy of 91% with the receiver operating characteristic (ROC) curves plotted in Fig. 4 (“CNN_FullData”), the corresponding area under the curve (AUC) was 0.9. Confusion matrix and performance report are presented in Tables 2 and 3, respectively.
Fig. 4.
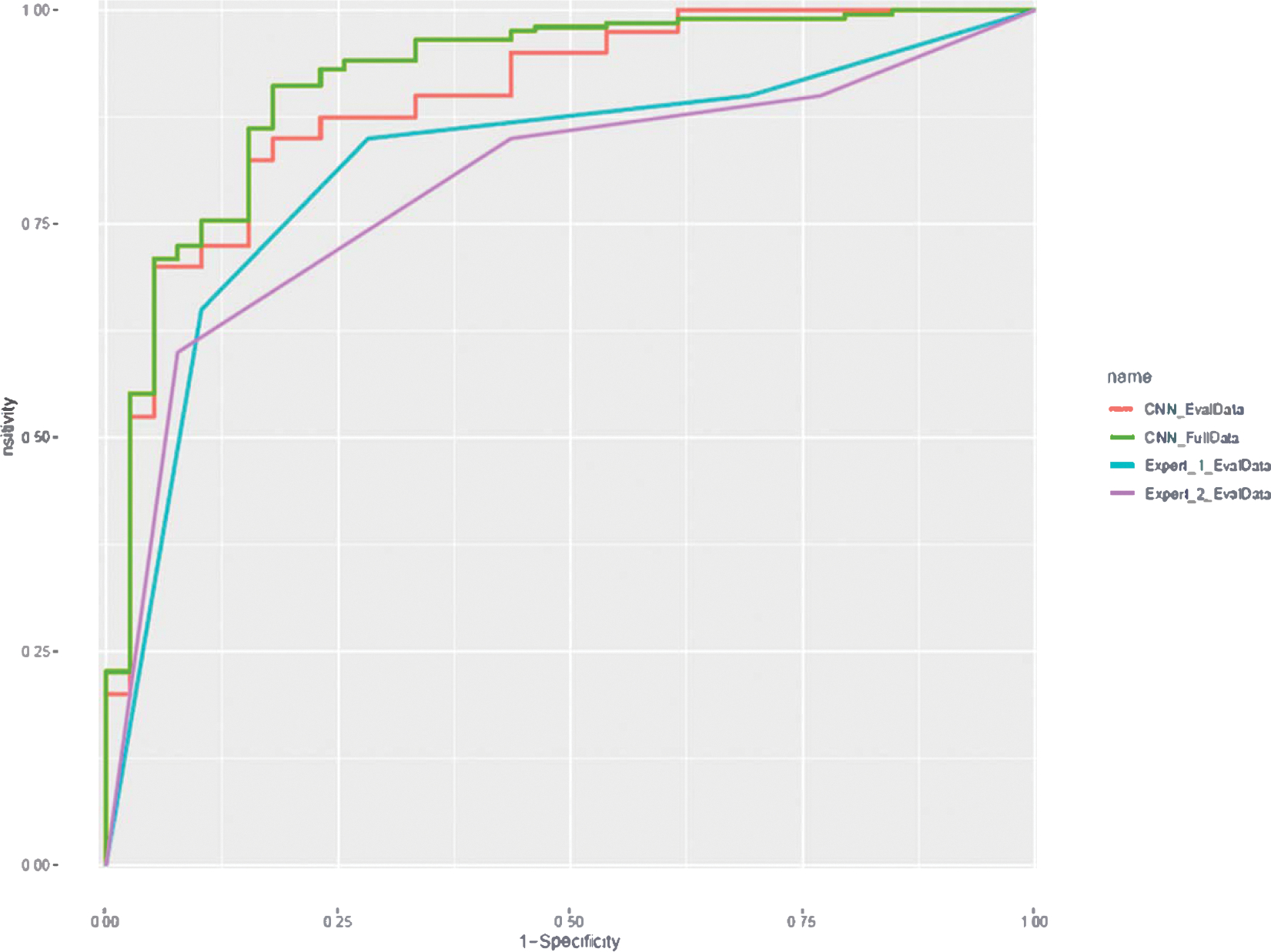
Receiver operating characteristic (ROC) curve plotted for performance of algorithm (blue for full data and orange for 80 lesion subset) for differentiating ccRCC from oncocytoma. Gray and yellow lines show radiologist performance on 80 lesion subset. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 2.
Confusion matrix for five-fold cross validation results in classification of ccRCC and oncocytoma lesions.
| Histology-based ground truth |
|||
|---|---|---|---|
| ccRCC | Oncocytoma | ||
|
| |||
| Prediction | ccRCC | 190 | 10 |
| Oncocytoma | 13 | 30 | |
Table 3.
Performance report for two radiologists and AI algorithm on comparison dataset (80 tumors) for classification of tumors to ccRCC vs. oncocytoma (binary decision).
| ccRCC | Sensitivity | Specificity | PPV | NPV | Accuracy |
|---|---|---|---|---|---|
| Radiologist 1 | 0.85 | 0.70 | 0.74 | 0.82 | 0.78 |
| Radiologist 2 | 0.85 | 0.55 | 0.65 | 0.79 | 0.70 |
| AI | 0.88 | 0.75 | 0.78 | 0.86 | 0.81 |
PPV: positive predictive value; NPV: negative predictive value.
Out of the 80 tumors that were reviewed by the radiologists, AI achieved the classification sensitivity of 88%, specificity of 75% and accuracy of 81% for predicting the ccRCC lesions, while the corresponding numbers were 85%, 70%, 78% for radiologist 1 and 85%, 55%, 70% for radiologist 2. The ROC curves are shown in Fig. 4 (*_EvalData), where the AI achieved an AUC of 0.9. Using radiologist 1–4 scoring system, each radiologist had in total 5 sensitivity-specificity pairs (including origin and [1, 1]): consider score 1/2/3/4 all as positive for ccRCC, consider score 2/3/4 as positive, consider score 3/4 as positive, consider score 4 as positive, and consider all scores negative. It could also be observed from the ROC curve that the subset ROC was roughly aligned with full ROC for CNN.
Among these 80 tumors reviewed by 2 radiologists, radiologist 1 selected 37 oncocytoma and 43 ccRCC. Radiologist 2 selected 35 oncocytoma and 45 ccRCC lesions. Intra-class correlation measurement resulted in a mean κ score of 0.56 for the classification task, indicating moderate agreements between the radiologists and AI.24 When certainty of decision making was considered, both radiologists and AI performed better for decisions made with high probability (>75%) than low probability (less than 75%). AI algorithm showed the highest performance, among three for high probability decisions and was correct in 94% of the times for ccRCC and 78% for oncocytoma tumors. These numbers were 89% and 75% for radiologist 1, and 86%, and 75% for radiologist 2, respectively. Moreover, AI also made higher number of decisions with high probability, i.e. 58 high probability decisions made by AI vs. 46 and 40 from the 2 radiologists. Performance decreased for all three for decisions made with low probability. Specifically, performance of AI had a significant decrease with an accuracy of 67% for ccRCCs and 69% for oncocytomas. These numbers were 80% and 67% for radiologist 1, and 83% and 46% for radiologist 2, respectively.
Out of 40 ccRCC lesions, 30 were correctly classified by both radiologists and AI. Fifteen ccRCC lesions were classified with high probability by all three entities. For oncocytomas, both radiologists and AI had agreement on correctly identifying 11 lesions. Two of these lesions were classified with high probability by all three entities. There were six oncocytoma lesions that were misclassified by radiologists for which AI had a correct classification. Table 4 shows the confusion matrix comparing performance of AI and radiologists on 80 tumor subset.
Table 4.
Shows confusion matrix for performance of the AI algorithm, radiologist 1, and radiologist 2; considering probability of decision making with decisions made with low certainty ( <75% - right) and high certainty (>75% - left).
| High certainty | Truth | Low certainty | Truth | ||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Rating | ccRCC | Oncocytoma | Rating | ccRCC | Oncocytoma | ||
|
|
|
||||||
| ccRCC | 29 | 6 | AI | ccRCC | 6 | 4 | AI |
| Oncocytoma | 2 | 21 | Oncocytoma | 3 | 9 | ||
|
|
|
||||||
| ccRCC | 26 | 4 | R d1 | ccRCC | 8 | 8 | R 1 |
|
|
|
||||||
| Oncocytoma | 4 | 12 | Oncocytoma | 2 | 16 | ||
|
|
|
||||||
| ccRCC | 24 | 3 | R d2 | ccRCC | 10 | 15 | R 2 |
|
|
|
||||||
| Oncocytoma | 4 | 9 | Oncocytoma | 2 | 13 | ||
4. Discussion
In the present study, we investigated whether ccRCC can be differentiated from oncocytoma lesions on pre and post-contrast MR imaging using AI based on deep learning with convolutional neural network. Our result indicates that AI can be a useful tool for differentiation of these two tumor subtypes with reasonable performance metrics.
It is evident that clinical outcomes of various types of renal masses are significantly different and successful application of a non-invasive method for risk stratification is of paramount importance, specifically in differentiation of ccRCC from oncocytoma lesions which has always been a diagnostic challenge for radiologists.25,26 This is particularly important for lesions that are considered small since these are followed with different active surveillance strategies. Various tools and imaging sequences have been previously tested for their ability in differentiating ccRCC from Oncocytoma.12,25,27,28 Most of these studies utilized imaging information from only a single classifier for this classification task. The advantage that AI algorithms could potentially bring about to this classification task is their ability in aggregating information from multiple tools and sequences. Coy et al evaluated the performance of Google TensorFlow™ in differentiation of ccRCC and oncocytoma on a cohort of 179 lesions. In their study, they utilized multiple phases of contrast-enhanced CT scans and reported an accuracy of 74.4%. However, their training method was limited by transfer learning from ImageNet datasets that contains non-medical imaging data. Additionally, the algorithm used in their study investigated individual performance of sequences for this prediction task, again utilizing only a single classifier for each performance measurement.29 As a preliminary effort in implementing such multiscale approach for differentiation of renal masses, we developed an in-house AI algorithm by utilizing a three-channel network by inputting combinations of four selected sequences of MR images. We selected these sequences in a way to take advantage of the information from contrast enhancement and fat and water tissue characteristics.
Our findings showed combining information from T2-WI and T1-post contrast sequences resulted in best performance of algorithm that could indicate the importance of utilizing information related to both T1 and T2-specific tissue characteristics and contrast phasing. Moreover, by selecting a fixed bounding box surrounding the tumor in the image-space, we also included the effect of perilesional information in our calculations. This could be important as renal tumors can cause physiologic changes in the surrounding tissue that might be specific to their unique pathology.30
To better understand our findings and outputs of AI, we asked two independent radiologists to classify a subset of 80 tumors which were also reviewed by the algorithm. However, instead of a binary classification task, we defined an observer study utilizing a rating system (1–4) to assess radiologists’ certainty of judgment. This way, we were able to generate probability of decision making from radiologists’ classifications, similar to the output from AI. Additionally, when considering probability of decision making, both radiologists and AI were more accurate for decisions made with higher confidence (probability >75%). This could indicate the strength of imaging metrics learned by the AI algorithm and radiologic evidence used by radiologists for differentiation of these two subtypes. Out of 40 ccRCC lesions, 15 were correctly identified by both radiologists and the algorithm with high probability. This may indicate that these tumors contained meaningful imaging patterns significantly suggestive of ccRCC pathology that were detected by deep network and radiologists with high certainty of classification. By reviewing these cases, we found that these lesions appeared to be heterogeneous cases of ccRCC that showed more enhancing septations on post-contrast studies with brighter appearance on T2 WI, same as the features previously reported in the literature.31 33 Fig. 5 shows an example of these tumors that showed significant evidence suggestive of ccRCC.
Fig. 5.

(A) T2-WI, (B) pre-contrast phase, (C) post-contrast arterial, and (D) post-contrast venous phase MRI image sets in a 34-year old male with a clear cell renal cell carcinoma diagnosed with von Hippel-Lindau syndrome. The mass was correctly identified by both radiologists and AI with high probability of being ccRCC.
Our algorithm had much better performance in identification of ccRCC tumors versus oncocytoma. We also observed that both radiologists performed better in identifying ccRCC tumors than oncocytoma lesions. Even though we used several measures of data augmentation to care for data imbalance, part of the reason for lower performance of the AI algorithm on oncocytomas could be due to smaller number of training samples of oncocytoma versus clear cell (40 vs. 203). However, our findings that both radiologists and AI had less accuracy in characterization of oncocytomas could emphasize the difficulty of differentiating oncocytoma lesions from ccRCCs potentially attributed to less unique imaging features characteristic of oncocytoma pathology. Out of 40 oncocytoma lesions, AI correctly classified 6 that were misclassified by both radiologists. These cases were rather smaller oncocytomas that showed enhancement patterns similar to ccRCC with heterogeneity and brighter appearance on T2-WI. Fig. 6 is an example of these oncocytoma tumors with features suggestive of ccRCC.
Fig. 6.
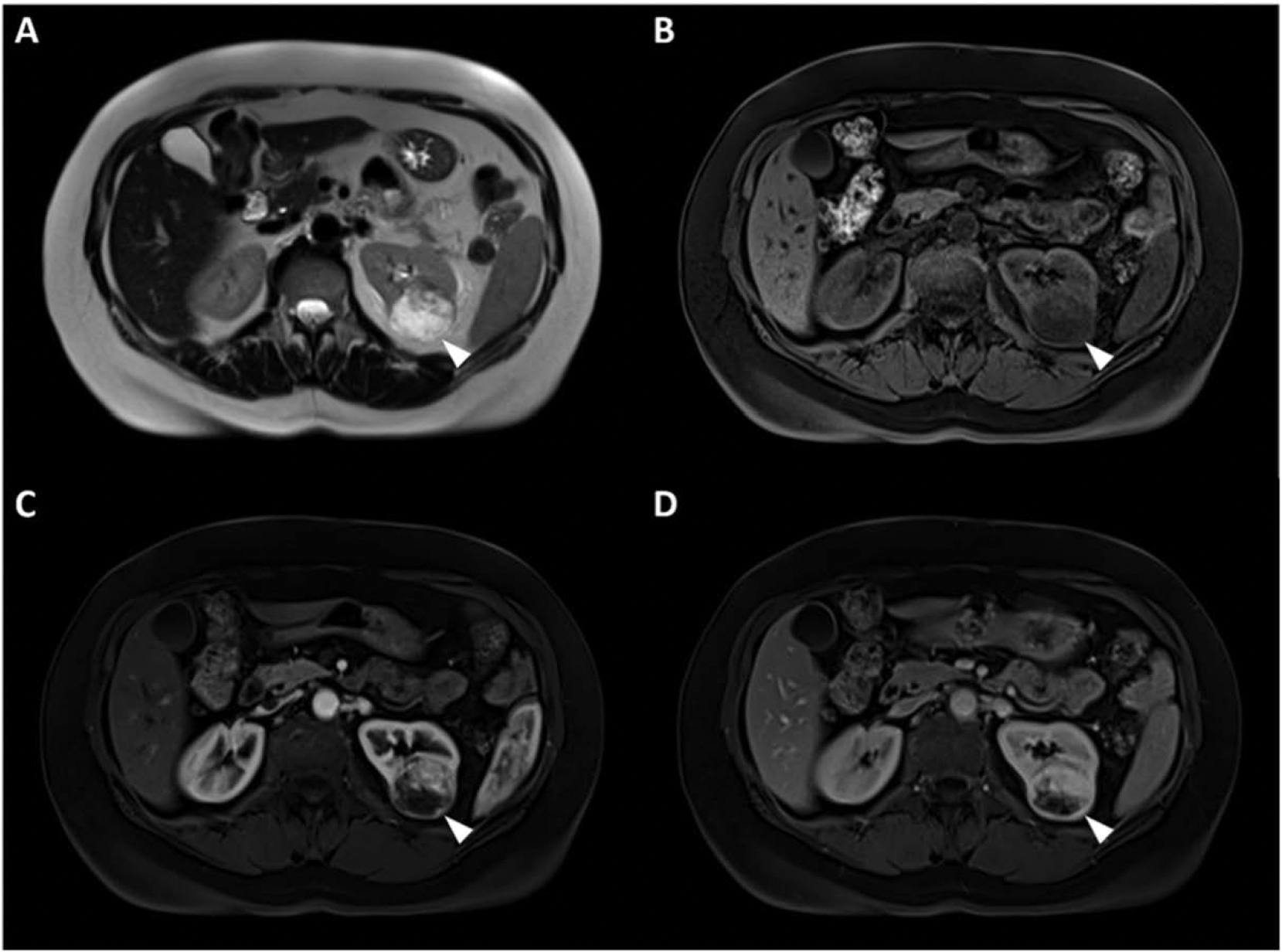
(A) T2 WI, (B) pre contrast phase, (C) post-contrast arterial, and (D) post contrast venous phase MRI image sets in a 41 year-old male with an oncocytoma tumor diagnosed with Birt-Hogg-Dubé syndrome. This mass was misclassified by both radiologists as ccRCC, but was correctly diagnosed by the AI algorithm. Imaging features of the mass represent common enhancement patterns similar to that of ccRCC.
The current study had some limitations. Even though we used a cross-validation method to address issues associated with overfitting, our findings are limited by the lack of a retest dataset. Another limitation was the small number of lesions used in this study particularly oncocytomas due to rare incidence of this subtype of renal tumors. Moreover, scores that were assigned by the radiologists were still subjective since there are no conventional radiologic guidelines for differentiation of these two tumor subtypes, ratings with lower probability would depend on expert judgment without sufficient clinical evidence. Additionally, our AI algorithm still needs a lesion to be detected and outlined for the classification task to be accomplished, however our group is working on building an AI system to automatedly detect kidney lesions, which will be cascaded to the defined lesion classification algorithm. Finally, at our institution, patients with kidney lesions are evaluated and managed by a dedicated radiology and urology team who has decades of experience in their respective fields. Thus, our single-institution study may not be readily generalizable to community settings.
5. Conclusion
In conclusion, this study showed that a deep learning based AI system using convolutional neural networks showed high diagnostic performance in differentiation of ccRCC and oncocytoma on multi-phasic MRIs.
Funding
This work was supported by the Intramural Research Programs of the Center for Cancer Research-National Cancer Institute and the National Institutes of Health Clinical Center, Bethesda, Maryland, USA. This research did not receive any specific grant from funding agencies in the public, commercial, or non-profit sectors.
Footnotes
Declaration of competing interest
None.
CRediT authorship contribution statement
Moozhan Nikpanah: Conceptualization, Investigation, Methodology, Visualization, Writing - Original draft preparation, Review and editing
Ziyue Xu: Methodology, Data curation, Formal analysis, Software, Writing - Original draft preparation, Review and editing
Dakai Jin: Methodology, Data curation, Formal analysis, Software, Writing - Original draft preparation, Review and editing
Faraz Farhadi: Conceptualization, Data curation, Investigation, Methodology, Visualization, Writing - Original draft preparation, Review & editing
Babak Saboury: Methodology, Writing - Review & editing
Mark W. Ball: Conceptualization, Writing - Review & editing
Rabindra Gautam: Methodology, Review & editing
Maria J. Merino: Investigation, Methodology, Review & editing
Bradford J. Wood: Methodology, Review & editing
Baris Turkbey: Methodology, Review & editing
Elizabeth C. Jones: Investigation, Methodology, Review & editing
W. Marston Linehan: Methodology, Funding acquisition, Resources, Validation, Review & editing
Ashkan A. Malayeri: Conceptualization, Funding acquisition, Investigation, Methodology, Resources, Supervision, Validation, Project administration, Writing - Review & editing.
References
- [1].Duchene DA, Lotan Y, Cadeddu JA, Sagalowsky AI, Koeneman KS. Histopathology of surgically managed renal tumors: analysis of a contemporary series. Urology 2003;62(5):827 30. [DOI] [PubMed] [Google Scholar]
- [2].Tsui KH, Shvarts 0, Smith RB, Figlin R, de Kemion JB, Belldegrun A. Renal cell carcinoma: prognostic significance of incidentally detected tumors. J Urol 2000; 163(2):426–30. [DOI] [PubMed] [Google Scholar]
- [3].Gill IS, Aron M, Gervais DA, Jewett MA. Small renal mass. N Engl J Med 2010;362(7):624–34. [DOI] [PubMed] [Google Scholar]
- [4].Capitanio U, Montorsi F. Renal cancer. Lancet 2016;387(10021):894 906. [DOI] [PubMed] [Google Scholar]
- [5].Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma 2017;3:17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sun MR, Ngo L, Genega EM, et al. Renal cell carcinoma: dynamic contrast-enhanced MR imaging for differentiation of tumor subtypes—correlation with pathologic findings 2009;250(3):793–802. [DOI] [PubMed] [Google Scholar]
- [7].Jinzaki M, Tanimoto A, Mukai M, et al. Double-phase helical CT of small renal parenchymal neoplasms: correlation with pathologic findings and tumor angiogenesis 2000;24(6):835–42. [DOI] [PubMed] [Google Scholar]
- [8].Bird VG, Kanagarajah P, Morillo G, et al. Differentiation of oncocytoma and renal cell carcinoma in small renal masses (< 4 cm): the role of 4 phase computerized tomography 2011;29(6):787 92. [DOI] [PubMed] [Google Scholar]
- [9].Alshumrani G, O’Malley M, Ghai S, et al. Small (≤ 4 cm) cortical renal tumors: characterization with multidetector CT 2010;35(4):488–93. [DOI] [PubMed] [Google Scholar]
- [10].Zhang J, Lefkowitz RA, Isbill NM, et al. Solid renal cortical tumors: differentiation with CT 2007;244(2):494–504. [DOI] [PubMed] [Google Scholar]
- [11].Cohen HT, McGovern FJ. Renalcell carcinoma. N Engl J Med 2005;353(23):2477 90. [DOI] [PubMed] [Google Scholar]
- [12].Young JR, Margolis D, Sauk S, Pantuck AJ, Sayre J, Raman SS. Clear cell renal cell carcinoma: discrimination from other renal cell carcinoma subtypes and oncocytoma at multiphasic multidetector CT. Radiology 2013;267(2):444 53. [DOI] [PubMed] [Google Scholar]
- [13].Role of percutaneous needle biopsy for renal masses. In: Caoili EM, Davenport MS, editors. Seminars in Interventional Radiology. Thieme Medical Publishers; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Phé V, Yates DR, Renard-Penna R, Cussenot O, Rouprêt M. Is there a contemporary role for percutaneous needle biopsy in the era of small renal masses? BJU Int 2012;109(6):867–72. [DOI] [PubMed] [Google Scholar]
- [15].Dechet CB, Bostwick DG, Blute ML, Bryant SC, Zincke H. Renal oncocytoma: multifocality, bilateralism, metachronous tumor development and coexistent renal cell carcinoma. J Urol 1999;162(1):40–2. [DOI] [PubMed] [Google Scholar]
- [16].Hoang UN, Mirmomen SM, Meirelles O, et al. Assessment of multiphasic contrast-enhanced MR textures in differentiating small renal mass subtypes 2018;43(12):3400–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Paschall AK, Mirmomen SM, Symons R, et al. Differentiating papillary type I RCC from clear cell RCC and oncocytoma: application of whole-lesion volumetric ADC measurement 2018;43(9):2424 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kocak B, Kus EA, Yardimci AH, Bektas CT, Kilickesmez O. Machine learning in radiomic renal mass characterization: fundamentals, applications, challenges, and future directions. Am J Roentgenol 2020:1–9. [DOI] [PubMed] [Google Scholar]
- [19].Sahiner B, Pezeshk A, Hadjiiski LM, et al. Deep learning in medical imaging and radiation therapy 2019;46(1):el 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Shin H-C, Roth HR, Gao M, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning 2016;35(5):1285 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yushkevich PA, Piven J, Hazlett HC, et al. User guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability 2006;31(3):1116–28. [DOI] [PubMed] [Google Scholar]
- [22].Imagenet classification with deep convolutional neural networks. In: Krizhevsky A, Sutskever I, Hinton GE, editors. Advances in Neural Information Processing Systems; 2012. [Google Scholar]
- [23].Caffe: convolutional architecture for fast feature embedding. In: Jia Y, Shelhamer E, Donahue J, Karayev S, Long J, Girshick R, et al. , editors. Proceedings of the 22nd ACM International Conference on Multimedia; 2014. [Google Scholar]
- [24].Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics 1977:159–74. [PubMed] [Google Scholar]
- [25].Chen F, Gulati M, Hwang D, et al. Voxelbased whole-lesion enhancement parameters: a study of its clinical value in differentiating clear cell renal cell carcinoma from renal oncocytoma 2017;42(2):552–60. [DOI] [PubMed] [Google Scholar]
- [26].Varghese BA, Chen F, Hwang DH, et al. Differentiation of predominantly solid enhancing lipid-poor renal cell masses by use of contrast enhanced CT: evaluating the role of texture in tumor subtyping 2018;211(6):W288 96. [DOI] [PubMed] [Google Scholar]
- [27].Yu H, Scalera J, Khalid M, et al. Texture analysis as a radiomic marker for differentiating renal tumors 2017;42(10):2470–8. [DOI] [PubMed] [Google Scholar]
- [28].Paño B, Macías N, Salvador R, et al. Usefulness of MDCT to differentiate between renal cell carcinoma and oncocytoma: development of a predictive model 2016;206(4):764–74. [DOI] [PubMed] [Google Scholar]
- [29].Coy H, Hsieh K, Wu W, et al. Deep learning and radiomics: the utility of Google TensorFlowTM Inception in classifying clear cell renal cell carcinoma and oncocytoma on multiphasic CT 2019;44(6):2009 20. [DOI] [PubMed] [Google Scholar]
- [30].Cao H, Fang L, Chen L, et al. The independent indicators for differentiating renal cell carcinoma from renal angiomyolipoma by contrast-enhanced ultrasound 2020; 20:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Rosenkrantz AB, Wehrli NE, Melamed J, Taneja SS, Shaikh MB. Renal masses measuring under 2 cm: pathologic outcomes and associations with MRI features. Eur J Radiol 2014;83(8):1311–6. [DOI] [PubMed] [Google Scholar]
- [32].Pedrosa I, Chou MT, Ngo L, et al. MR classification of renal masses with pathologic correlation 2008;18(2):365 75. [DOI] [PubMed] [Google Scholar]
- [33].Pedrosa I, Alsop DC, Rofsky NM. Magnetic resonance imaging as a biomarker in renal cell carcinoma. Cancer 2009;115(S10):2334–45. [DOI] [PubMed] [Google Scholar]


