Fig. 6. shRNA-mediated knockdown of E(z) and ash1 restores normal eyes development in respectively H3.3K36M and H3.3K27M mutant flies.
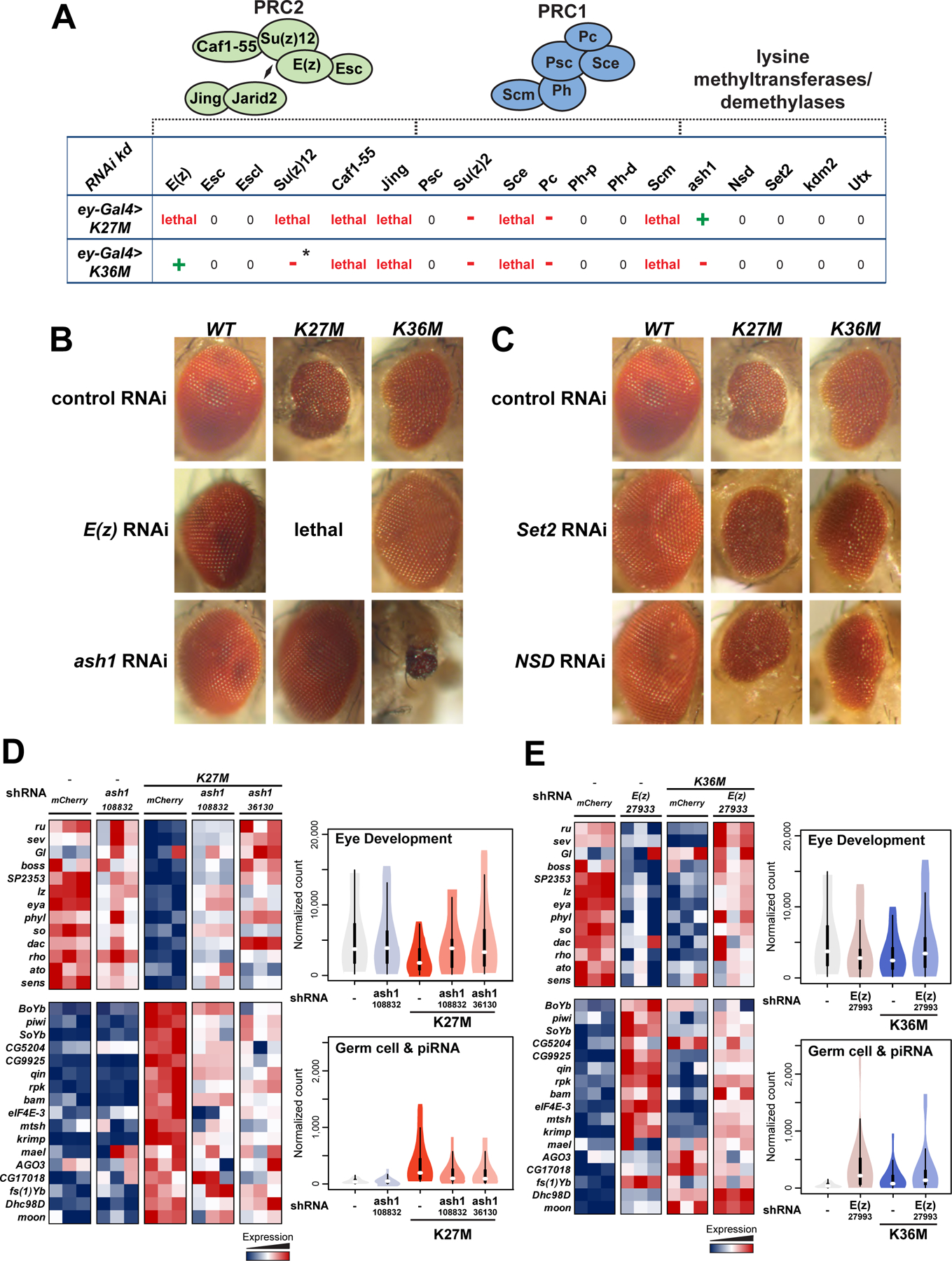
(A) Eye disc-specific H3.3K27M and H3.3K36M expressing flies were crossed with 18 selected strains expressing specific short hairpin (sh)RNAs targeting PRC2/PRC1 members as well as lysine-methyltransferases or demethylases. Green crosses indicate suppression of phenotype, red dashes indicate worsening of phenotype, while zeroes indicate no effect. Asterisk indicates that the effect was only observed in females, not in males. (B) Micrographs of adult eyes illustrating modifier effects of E(z) (BDSC: 27933) and ash1 (VDRC:108832) knockdown on ey>H3.3 WT, ey>H3.3K27M, and ey>H3.3K36M flies. (C) Micrographs of adult eyes illustrating the lack of a modifier effect from Set2 or NSD knockdown on ey>H3.3 WT, ey>H3.3K27M, and ey>H3.3K36M flies. (D) Left: heatmap depicting relative expression of eye developmental genes (upper panel) and germ cell genes (lower panel) in ash1-KD rescued H3.3K27M, relative to ash1-KD alone and mCherry KD controls. Right: violin plot representing expression quantification of gene sets depicted in the heatmap. (E) Left: heatmap depicting relative expression of eye developmental genes (upper panel) and germ cell genes (lower panel) in E(z)-KD rescued H3.3K36M, relative to E(z)-KD alone and mCherry KD controls. Right: violin plot representing expression quantification of gene sets depicted in the heatmap. See also Fig. S7, Table S6–9.
