Abstract
RNA exosome is a highly conserved ribonuclease complex essential for RNA processing and degradation. Bi-allelic variants in exosome subunits EXOSC3, EXOSC8 and EXOSC9 have been reported to cause pontocerebellar hypoplasia type 1B, type 1C and type 1D, respectively, while those in EXOSC2 cause short stature, hearing loss, retinitis pigmentosa and distinctive facies. We ascertained an 8-months-old male with developmental delay, microcephaly, subtle dysmorphism and hypotonia. Pontocerebellar hypoplasia and delayed myelination were noted on neuroimaging. A similarly affected elder sibling succumbed at the age of 4-years 6-months. Chromosomal microarray returned normal results. Exome sequencing revealed a homozygous missense variant, c.104C > T p.(Ser35Leu) in EXOSC1 (NM_016046.5) as the possible candidate. In silico mutagenesis revealed loss of a polar contact with neighboring Leu37 residue. Quantitative real-time PCR indicated no appreciable differences in EXOSC1 transcript levels. Immunoblotting and blue native PAGE revealed reduction in the EXOSC1 protein levels and EXO9 complex in the proband, respectively. We herein report an individual with the bi-allelic variant c.104C>T p.(Ser35Leu) in EXOSC1 and clinical features of pontocerebellar hypoplasia type 1. Immunoblotting and blue native PAGE provide evidence for the pathogenicity of the variant. Thus, we propose EXOSC1 as a novel candidate gene for pontocerebellar hypoplasia.
1. INTRODUCTION
The RNA exosome is an evolutionarily conserved and ubiquitously expressed ribonuclease complex. It is essential for the surveillance, processing, and degradation of various classes of RNA in the nucleus and cytoplasm with 3′- 5′ exoribonuclease and endonuclease activity1-3. The cytoplasmic exosome complex consists of 10 subunits. Six of these proteins, EXOSC4 to EXOSC9, harboring a “RNase PH-like domain”, form the barrel-shaped central core. Three proteins, EXOSC1, EXOSC2 and EXOSC3, form the cap and aid in stable assembly of the core. These subunits consist of an N-terminal domain and S1-KH RNA-binding domains4, 5. These nine subunits (collectively called EXO9) are catalytically inactive and form the structural component of RNA exosomes. DIS3 (EXOSC11), forms the catalytic component of the complex and associates at the base of the central core. RNAs are guided down through the EXO9 core to the DIS3 for processing4, 6-8.
Pathogenic variants in exosome complex proteins, EXOSC3, EXOSC8, EXOSC9, EXOSC2 and its cofactors, RBM7, SKIV2L, TTC37 are reported to cause human diseases9-15. Three of the four subunits complex proteins, (EXOSC3, EXOSC8 and EXOSC9) are associated with subtypes of pontocerebellar hypoplasia type 19, 10, 15. Defects in the fourth subunit, encoded by EXOSC2, cause a distinct syndrome of short stature, hearing loss, retinitis pigmentosa and distinctive facies (MIM# 617763). However, progressive cerebellar atrophy is a component of this syndrome as well11. In this study, we report a child with developmental delay, microcephaly, facial dysmorphism, short stature, hypotonia, pontocerebellar hypoplasia and delayed myelination due to a missense variant, c.104C > T in EXOSC1.
2. MATERIALS AND METHODS
2.1. Clinical report
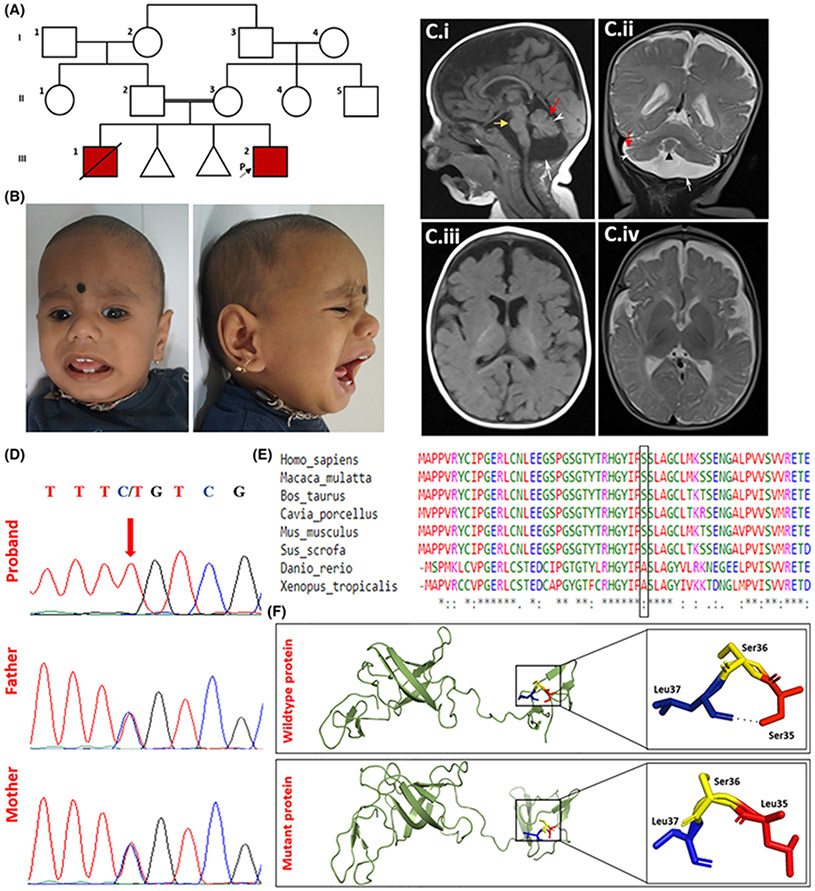
We ascertained an 8-months-old male with developmental delay. He is the second born to a third-degree consanguineous couple (Figure 1A,B). On examination, blue sclera, tall forehead, telecanthus, strabismus, depressed nasal bridge, anteverted nares, thick vermillion border of lips, long and smooth philtrum and retrognathia were noted (Figure 1B). Magnetic resonance imaging (MRI) of the brain at 8 months of age showed hypoplastic cerebellum with mild dilatation of folia, vermian hypoplasia, relative sparing of pons, prominent prepontine and cerebellopontine angle cisterns. A prominent cerebrospinal fluid intensity was noted in the posterior fossa with mild elevation of the cerebellum, likely to be mega cisterna magna. Thinning of corpus callosum and mild delay in myelination was noted (Figure 1C). There is also prominent extra-axial fluid space in the frontotemporal region suggestive of associated cerebral atrophy. Detailed clinical findings are provided in the supplementary information.
FIGURE 1.
Clinical, radiological, molecular and in silico findings in the proband. A, Pedigree of the family. B, Clinical photographs of the proband. C, MRI of the brain at 8 months shows cerebellar hypoplasia (white arrowhead), mega cisterna magna (white arrow), mild dilatation of cerebellar folia (red arrow), vermian hypoplasia (black arrowhead), relatively preserved pons (yellow arrow) (i and ii), mild hypoplasia of corpus callosum. (i) and mild delayed myelination according to age (iii and iv). Extra-axial CSF spaces in bilateral frontotemporal region appear prominent (iv) suggestive of associated cerebral atrophy. D, Electropherograms of the sequence variant, c.104C > T in EXOSC1 in the proband (homozygous) and parents (heterozygous). E, Multiple alignment of EXOSC1 sequences across multiple species (Ser35 highlighted). F, Protein structure prediction shows replacement of the Ser35 with mutant Leu35 is predicted to lead to loss of a polar contact resulting in altered stability of the protein.
2.2. Genetic analysis and functional validation
Details of genetic testing, in silico protein modeling, cell culture, RNA extraction and RT-PCR, quantitative real-time PCR (qRT-PCR), immunoblotting, Blue native PAGE are provided in the supplementary information.
3. RESULTS
3.1. Variant identification
Chromosomal microarray returned normal results. Exome sequencing revealed a missense variant, c.104C>T p.Ser35Lue in the exon 2 of EXOSC1 (NM_016046.5) in homozygous state as the possible candidate. His parents were found to be heterozygous carriers of the variant (Figure 1D). This variant was not observed in gnomAD and our in-house exome data of 960 individuals. The variant lies in the N-terminal RPL27-like domain of EXOSC1 protein. The amino acid at this position is highly conserved across mammals (GERP_RS score: 5.44). In silico tools such as MutationTaster, Polyphen-2, SIFT, M-CAP, CADD, FATHMM_MKL and REVEL predicted the variant to be damaging to the protein function.
3.2. Functional validation
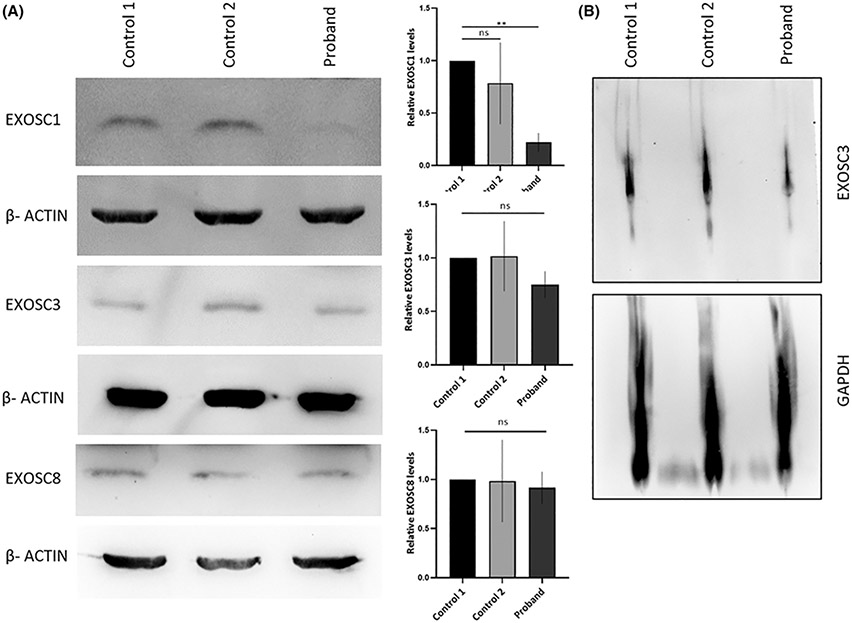
Upon sequence homology-based protein modeling of EXOSC1, the wild-type amino acid residue, Ser35 was noted to have a single polar contact with neighboring residue Leu37. In silico mutagenesis of wild-type Ser35 to mutant Leu35 resulted in loss of the said polar interaction. This is likely to result in altered secondary protein structure of EXOSC1 (Figure 1F). RT-PCR detected no abnormal splicing due to the presence of the variant, c.104C>T in EXOSC1, and qRT-PCR showed no change in the expression of ESOXC1 mRNA in the proband when compared to two unrelated healthy controls (Supplementary Figure 1). Immunoblotting performed on cultured fibroblasts lysate to detect the expression of RNA exosome complex proteins, EXOSC1, EXOSC3 and EXOSC8 revealed a significant reduction in the EXOSC1 protein in the proband compared with two healthy controls. EXOSC3 and EXOSC8 protein levels remained unaltered (Figure 2A). BN-PAGE performed using protein lysate from the fibroblasts of proband and two healthy controls revealed the significant reduction of the exosome complex in the proband (Figure 2B).
FIGURE 2.
Immunoblotting and BN-PAGE determine reduced levels of EXOSC1 and exosome complex. Immunoblotting of RNA exosome complex proteins, EXOSC1, EXOSC3 and EXOSC8 in patient and controls. Significant reduction in EXOSC1 was observed in the proband compared to two unrelated healthy controls. EXOSC3 and EXOSC8 levels remained unaltered in the patient. β-Actin was used as loading control. Results are the mean ± SEM of at least three different experiments (**P < .05, One-way ANOVA followed by Dunnett's multiple comparison test). B, BN-PAGE revealed a reduction of the assembly of the exosome complex in the proband's fibroblast. GAPDH was used as a loading control.
4. DISCUSSION
The proband presented with clinical features consistent with PCH type 1 and a bi-allelic missense variant, c.104C>T in EXOSC1. The phenotype is similar to defects in the closely related proteins of the exosome complex. Immunoblotting and blue native PAGE experiments with patient's fibroblast provide functional evidence for the pathogenicity of the variant.
Individuals with pathogenic variants in EXOSC3, EXOSC8 and EXOSC9 present with developmental delay, failure to thrive, recurrent pulmonary infections, hypotonia progressing to spasticity, cerebellar signs like nystagmus and tremors, subtle facial dysmorphism with or without seizures and early demise. Muscle weakness secondary to central and peripheral motor dysfunction has been noted in all PCH type 1 subtypes. Nerve conduction studies showed presence of axonal neuropathy and muscle biopsy revealed the findings of spinal muscular atrophy (SMA) like muscle atrophy in most. Although the proband has significant axial and peripheral hypotonia, these tests could not be performed. Regression of attained milestones have been noted to occur post seizure episodes. Of note, blue sclera was noted in the current proband and individuals affected with EXOSC9-related PCH. Other systemic findings include hearing and vision impairment, oculomotor apraxia, retinal dystrophy, cholelithiasis and tongue atrophy and fasciculations, hyperthyroidism, glaucoma, retinitis pigmentosa, corneal dystrophy and atrial hypertension9, 10, 15. MRI brain in the present proband showed hypoplasia of cerebellum and vermis with relative sparing of pons, thinning of corpus callosum and delayed myelination. These findings are similar to those observed in PCH type 116. Phenotypic and genotypic comparison of exosome complex related disorders are provided in the Table 1.
TABLE 1.
Phenotypic and genotypic comparison of exosome complex-related disorders
| Gene | EXOSC1 | EXOSC8 | EXOSC9 | EXOSC3 | EXOSC2 |
|---|---|---|---|---|---|
| Disorder | Pontocerebellar hypoplasia (proband) | Pontocereblellar hypoplasia type 1C | Pontocereblellar hypoplasia type 1D | Pontocerebellar hypoplasia, type 1B | Short stature, hearing loss, retinitis pigmentosa and distinctive facies |
| Number of families (individuals) | Not applicable | 3 (22) | 7 (7) | 58 (82) | 2 (3) |
| Type of sequence variant | Missense | Missense | Missense, stopgain in trans with missense | Missense, frameshift deletion, splicing variant | Missense |
| Clinical findings | |||||
| Age of onset | Infancy | Infancy | Intrauterine life to infancy | Infancy | Variable (birth to third decade) |
| Developmental delay | Yes | Yes | Yes | Yes | Yes |
| Feeding difficulties | No | No | Yes (in few) | Yes (in few) | No |
| Seizures | No | No | Yes (in two related individuals) | Yes (in few) | No |
| Growth retardation | Yes | Yes | Yes | Yes | Yes |
| Facial dysmorphism | Yes | Yes (in some) | Yes (in some) | Yes | Yes |
| Microcephaly | Yes | Yes | Yes | Yes | Yes |
| Tone | Hypotonia | Spastic tetraparesis | Severe hypotonia of limbs | Axial hypotonia to spasticity | NA |
| Deep tendon reflexes | Diminished | Diminished/exaggerated | Absent | Diminished (in few) | NA |
| MRI findings | |||||
| Cerebellar signs | No | Tremors | Nystagmus | Intention tremors, nystagmus | Nystagmus |
| Joint contractures | No | Yes | Yes (arthrogryposis multiplex congenita) | Yes | NA |
| Cerebellar hemisphere hypoplasia | Yes | Yes | Yes | Yes | Yes |
| Cerebellar vermian hypoplasia | Yes | Yes | Yes | Yes | No |
| Dragonfly appearance | No | No | No | Yes (in few) | No |
| Dilatation of cerebellar folia | Yes | No | No | No | No |
| Cerebellar cysts | No | No | No | Yes | No |
| Pontine hypoplasia | Yes | Yes (in few) | Yes (in few) | Yes (in few) | No |
| Cerebral atrophy | Yes | Yes | Yes (in few) | Yes | No |
| Corpus callosum abnormalities | Thinning of corpus callosum | Thinning of corpus callosum | No | No | No |
| Myelination abnormalities | No | Yes | No | No | Yes |
| Other investigations | |||||
| Muscle biopsy | Not done | SMA-like findings | SMA-like findings | SMA-like findings | NA |
| NCV/EMG | Not done | Motor neuronopathy | Axonal motor neuronopathy | Neurogenic changes on EMG | NA |
| Ophthalmology evaluation | Blue sclera | Impaired vision | Blue sclera, absent fixation, impaired pursuit | Oculomotor apraxia, poor visual attention, nystagmus, strabismus, retinal dystrophy | Retinitis pigmentosa, corneal dystrophy, glaucoma, nystagmus, strabismus |
| Hearing evaluation | Not done | SNHL | Normal (available for four probands) | Not available | SNHL |
Abbreviations: EMG, electromyography; MRI, magnetic resonance imaging; NCV, nerve conduction velocity study; NA, not available; SMA, spinal muscular atrophy; SNHL, sensory neural hearing loss.
EXOSC1 encodes a structural component of the RNA exosome complex that is involved in the general processing and degradation of coding and non-coding RNAs4. The variant identified in this study, p.Ser35Lue is present in the highly conserved N-terminal RPL27-like domain of EXOSC1. Transcript analysis followed by immunoblotting in the patient fibroblast revealed that even with the normal EXOSC1 mRNA expression the protein was significantly reduced. This finding suggests that change in the amino acid from serine to leucine at p.35 might affect the stability of the EXOSC1 protein. Previously, missense variants in different exosome subunits were shown to significantly reduce the amount of protein9, 10, 17. Each subunit of the complex is critical for its assembly and defects in any one subunit might affect the complex formation10. Previous studies in individuals harboring missense variants in three exosome subunits, showed a consequent reduction in the exosome complex assembled9, 10, 17. Similarly we observed that the EXOSC1 variant (p.Ser35Leu) resulted in a decrease in the exosome complex detected in proband fibroblasts, although this was less marked compared to that seen previously in EXOSC3 and EXOSC8 mutant primary cells. This supports the notion that the primary reduction in the levels of any one component of the exosome destabilizes the complex assembly.
Studies using model organisms have revealed the critical role of the exosome complex in brain development. Knockdown experiments of exosc9 in zebrafish, showed defect in development of cerebellum and hindbrain hypoplasia, and motor neurons migration10. Poor motility and small brain size were observed in exosc3 knockdown zebrafish models15. Downregulation exosc8 in zebrafish showed abnormal midbrain, hindbrain and impaired myelination9. Cerebellar atrophy and microcephaly were observed in exosc8 and exosc9 mutant zebrafish with significant increase in the transcript levels of p53. Increased p53 correlates with increased number of brain cells undergoing apoptosis during development17. These findings reflect phenotypes observed in the patients with variants in genes coding for exosome subunits and the importance of exosome complex in brain development.
In this study, we propose EXOSC1 as a novel candidate gene for pontocerebellar hypoplasia type 1. Report of additional individuals and further studies in model organisms would confirm EXOSC1 as a cause of pontocerebellar hypoplasia EXOSC1 related pontocerebellar hypoplasia.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to patient and his family for participating in the study. We thank Dr Govindaraj Periyasamy, Institute of Bioinformatics, Bengaluru and Deepha Sekar, Department of Neuropathology, NIMHANS, Bengaluru for their valuable suggestions in performing BN-PAGE experiment. This work was funded by Department of Health Research, Ministry of Health and Family Welfare, Government of India under the project titled “Clinical and Molecular Characterization of Leukodystrophies in Indian Children” grant ID—V.25011/379/2015-GIA/HR and National Institutes of Health, USA under the project titled ‘Genetic Diagnosis of Neurodevelopmental Disorders in India’ Grant ID—R01 HD093570 01 A1 (PI).
Footnotes
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from corresponding author upon reasonable request.
REFERENCES
- 1.Chlebowski A, Lubas M, Jensen TH, Dziembowski A. RNA decay machines: the exosome. Biochim Biophys Acta. 2013; 1829(6–7): 552–560. [DOI] [PubMed] [Google Scholar]
- 2.Januszyk K, Lima CD. The eukaryotic RNA exosome. Curr Opin Struct Biol. 2014; 24: 132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eberle AB, Visa N. Quality control of mRNP biogenesis: networking at the transcription site. Semin Cell Dev Biol. 2014; 32: 37–46. [DOI] [PubMed] [Google Scholar]
- 4.Morton DJ, Kuiper EG, Jones SK, Leung SW, Corbett AH, Fasken MB. The RNA exosome and RNA exosome-linked disease. RNA. 2018; 24(2): 127–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmid M, Jensen TH. The nuclear RNA exosome and its cofactors. Adv Exp Med Biol. 2019; 1203: 113–132. [DOI] [PubMed] [Google Scholar]
- 6.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006; 127(6): 1223–1237. [DOI] [PubMed] [Google Scholar]
- 7.Makino DL, Baumgärtner M, Conti E. Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature. 2013; 495(7439): 70–75. [DOI] [PubMed] [Google Scholar]
- 8.Malet H, Topf M, Clare DK, et al. RNA channelling by the eukaryotic exosome. EMBO Rep. 2010; 11(12): 936–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boczonadi V, Müller JS, Pyle A, et al. EXOSC8 mutations alter mRNA metabolism and cause hypomyelination with spinal muscular atrophy and cerebellar hypoplasia. Nat Commun. 2014; 5: 4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burns DT, Donkervoort S, Müller JS, et al. Variants in EXOSC9 disrupt the RNA exosome and result in cerebellar atrophy with spinal motor neuronopathy. Am J Hum Genet. 2018; 102(5): 858–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Donato N, Neuhann T, Kahlert A-K, et al. Mutations in EXOSC2 are associated with a novel syndrome characterised by retinitis pigmentosa, progressive hearing loss, premature ageing, short stature, mild intellectual disability and distinctive gestalt. J Med Genet. 2016; 53(6): 419–425. [DOI] [PubMed] [Google Scholar]
- 12.Fabre A, Charroux B, Martinez-Vinson C, et al. SKIV2L mutations cause syndromic diarrhea, or trichohepatoenteric syndrome. Am J Hum Genet. 2012; 90(4): 689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabre A, Martinez-Vinson C, Roquelaure B, et al. Novel mutations in TTC37 associated with tricho-hepato-enteric syndrome. Hum Mutat. 2011; 32(3): 277–281. [DOI] [PubMed] [Google Scholar]
- 14.Hartley JL, Zachos NC, Dawood B, et al. Mutations in TTC37 cause trichohepatoenteric syndrome (phenotypic diarrhea of infancy). Gastroenterology. 2010; 138(7): 2388–2398.e1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wan J, Yourshaw M, Mamsa H, et al. Mutations in the RNA exosome component gene EXOSC3 cause pontocerebellar hypoplasia and spinal motor neuron degeneration. Nat Genet. 2012; 44(6): 704–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivanov I, Atkinson D, Litvinenko I, et al. Pontocerebellar hypoplasia type 1 for the neuropediatrician: genotype–phenotype correlations and diagnostic guidelines based on new cases and overview of the literature. Eur J Paediatr Neurol. 2018; 22(4): 674–681. [DOI] [PubMed] [Google Scholar]
- 17.Müller JS, Burns DT, Griffin H, et al. RNA exosome mutations in pontocerebellar hypoplasia alter ribosome biogenesis and p53 levels. Life Sci Alliance. 2020; 3(8): e202000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from corresponding author upon reasonable request.