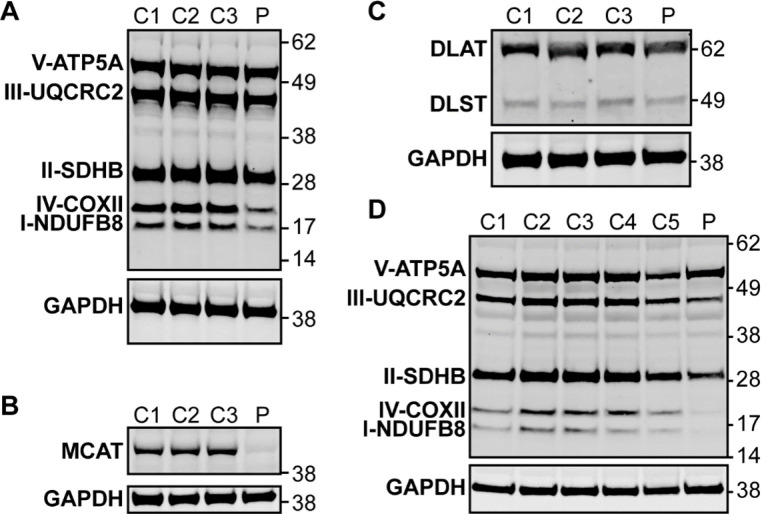
Figure 2. Western blot analysis in patient lymphoblasts (A–C) and fibroblasts (D).
(A) Western blot analysis to assess the expression level of one subunit of each of the five different oxidative phosphorylation complexes in the patient lymphoblast sample (P) compared to three control lymphoblast lines from healthy individuals (C1–3). Protein content of NDUFB8 and COXII, which are subunits of complexes I and IV, respectively, are decreased in the patient compared to healthy controls. (B) Western blot analysis to assess malonyl-CoA-acyl carrier protein transacylase (MCAT) levels reveals decreased expression of MCAT in patient lymphoblasts (P) compared to three controls (C1–3). (C) Western blot analysis to assess lipoylation with an anti-lipoic acid antibody in patient lymphoblasts (P) compared to controls (C1–3) reveals normal lipoylation of the PDH and OGDH E2 components (DLAT and DLST, respectively) in the patient sample. (D) Western blot analysis to assess the expression level of one subunit of each of the five different oxidative phosphorylation complexes in the patient fibroblast sample (P) compared to five fibroblast controls (C1–5). Protein content of NDUFB8, COXII, and SDHB, which are subunits of complexes I, IV, and II, respectively, are decreased in the patient compared to healthy controls.
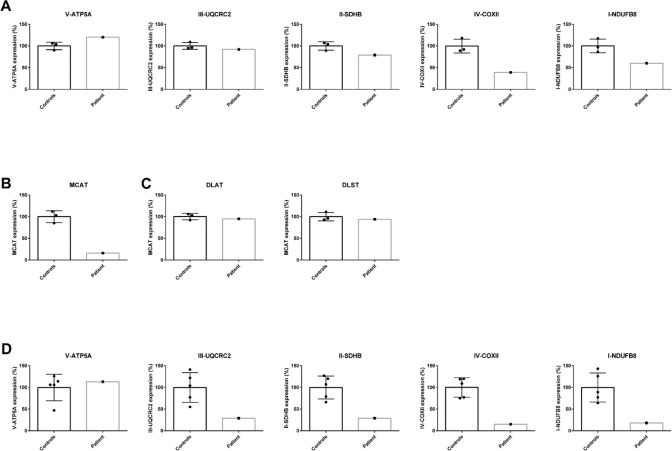
Figure 2—figure supplement 1. Quantification of western blot analysis in patient lymphoblasts (A–C) and fibroblasts (D).
Figure 2—figure supplement 2. Measurement of 2-oxoglutaric acid dehydrogenase complex (OGDHc) activity.
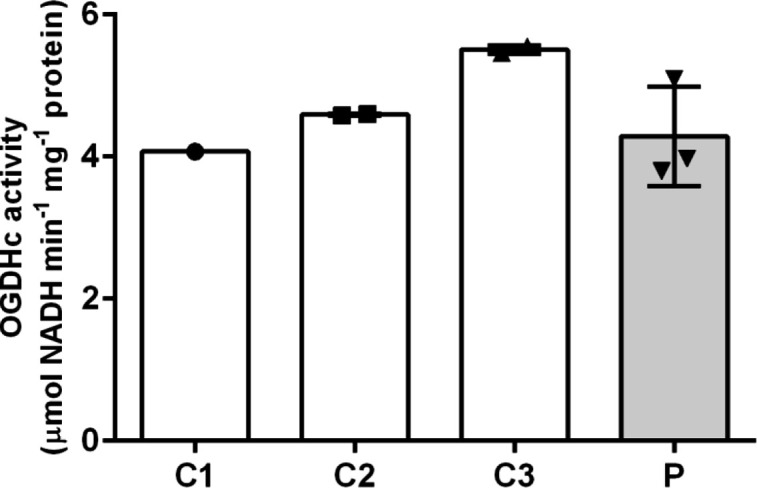
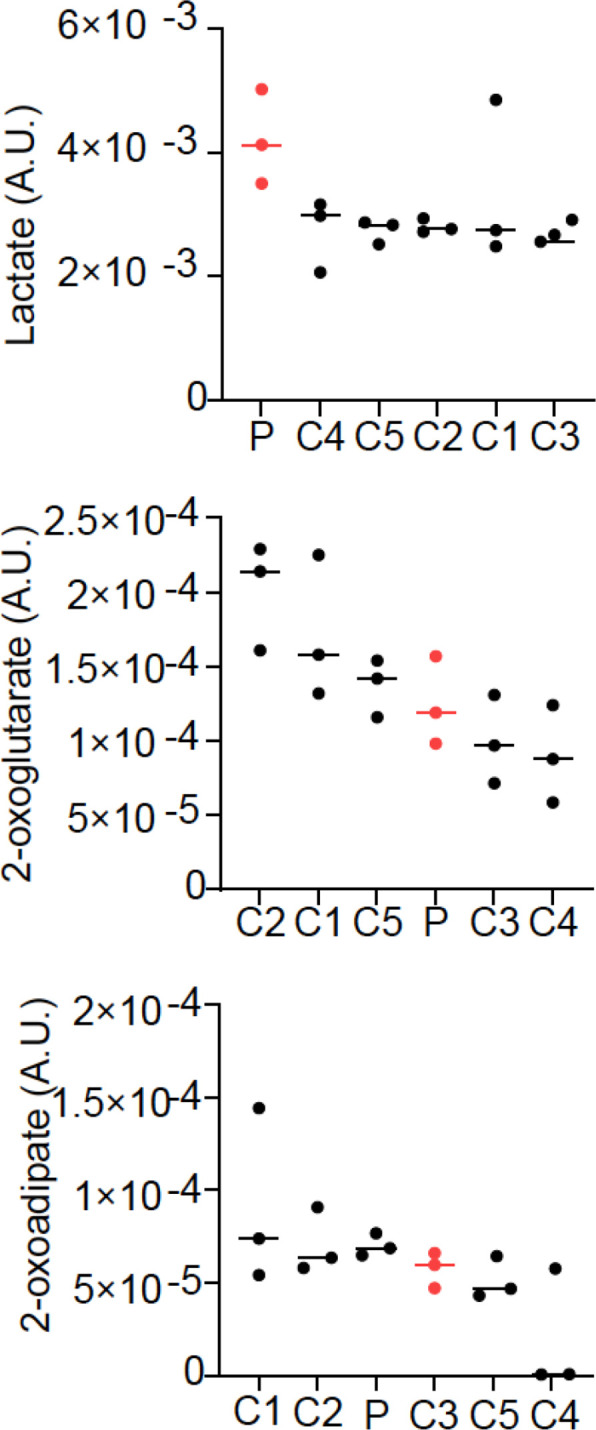
Figure 2—figure supplement 3. Relative abundance of lactate, 2-oxoglutarate, and 2-oxoadipate from quantitative metabolomics.