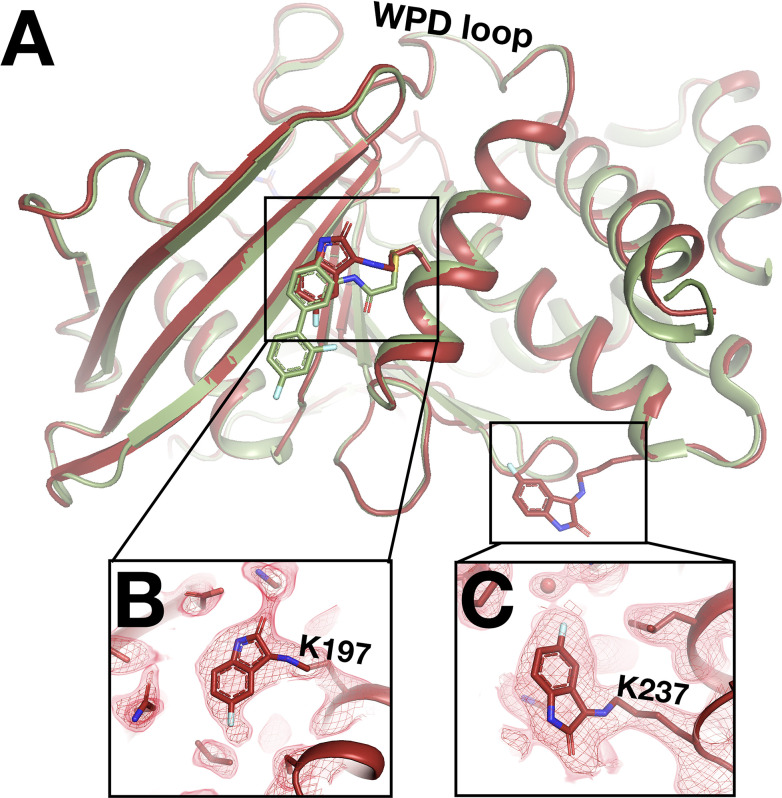
Figure 9. Unanticipated covalent adducts at previously reported allosteric sites only at room temperature (RT).
(A) RT structure with the fragment covalently bound to both K197 and K237 (z0048, red), aligned with cryogenic (cryo) structure with a previously reported allosteric inhibitor covalently bound to K197C (6b95, green). (B) Fragment bound to K197 at the 197 allosteric site, with RT event density at 1.5 σ. (C) Fragment bound to K237 at the L16 allosteric site, with RT event density at 1.5 σ.