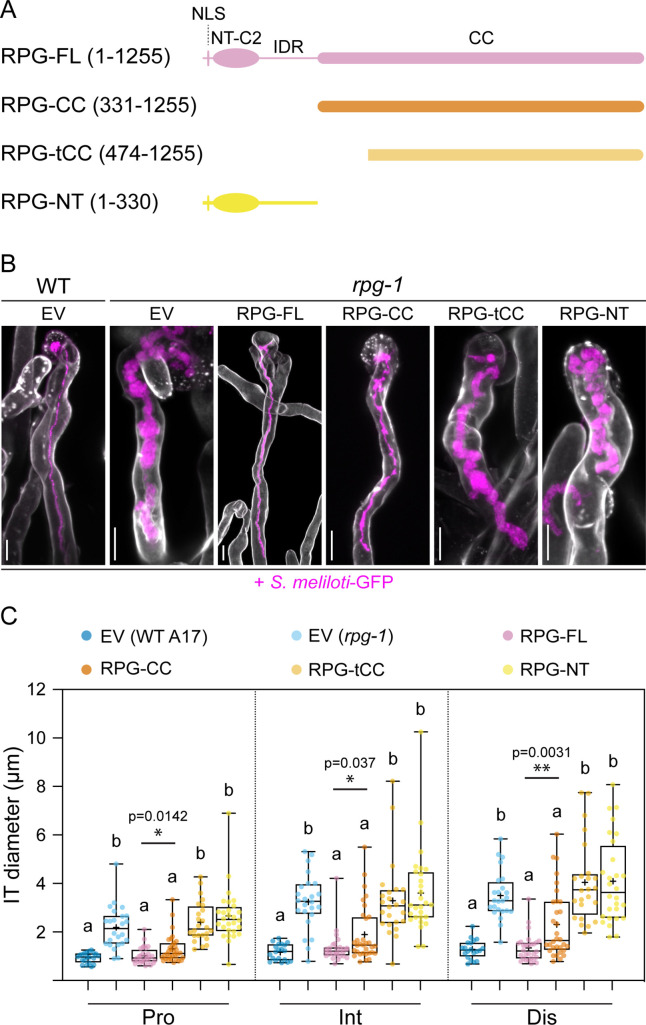
Figure 2. The coiled-coil domain of Rhizobium-directed polar growth (RPG) is necessary to restore WT-like infection thread morphology in rpg-1 root hairs.
(A) Schematic representation of the RPG full-length protein and deletion derivatives used for complementation assays. Numbers indicate amino acids included in each fragment. NLS = nuclear localization signal; NT-C2=N-terminal C2 domain; IDR = intrinsically disordered region; CC = coiled coil domain. (B) Representative confocal images of infection threads (ITs) developed within root hairs of wild-type (WT) and rpg-1 transgenic roots expressing the different constructs 21 dpi with S. meliloti-GFP (magenta). Cell walls were stained with Calcofluor white (white). Images are merges of maximum-intensity projections of fluorescent channels. EV = empty vector. Scale bars = 10 µm. (C) IT diameters scored at the three different points schematically represented in Figure 1C. In the box plot, the top and bottom of each box represents the 75th and 25th percentiles, the middle horizontal bars indicate the median and the whiskers represent the range of minimum and maximum values. Crosses represent sample means. Letters indicate statistically significant differences according to Kruskal-Wallis multiple comparison analysis followed by a Dunn’s post-hoc test. A Mann-Whitney test was performed to compare RPG-FL and RPG-CC with p-values <0.05 (*), <0.01 (**), and <0.001 (***). Data are from one of two independent experiments showing similar tendencies, with five (EV in WT), six (EV in rpg-1), seven (RPG-FL,), eight (RPG-CC), six (RPG-tCC), seven (RPG-NT) composite plants analyzed (one root per plant) and four ITs per roots. n=20 (EV in WT), 24 (EV in rpg-1), 28 (RPG-FL), 32 (RPG-CC), 24 (RPG-tCC), 28 (RPG-NT) ITs.