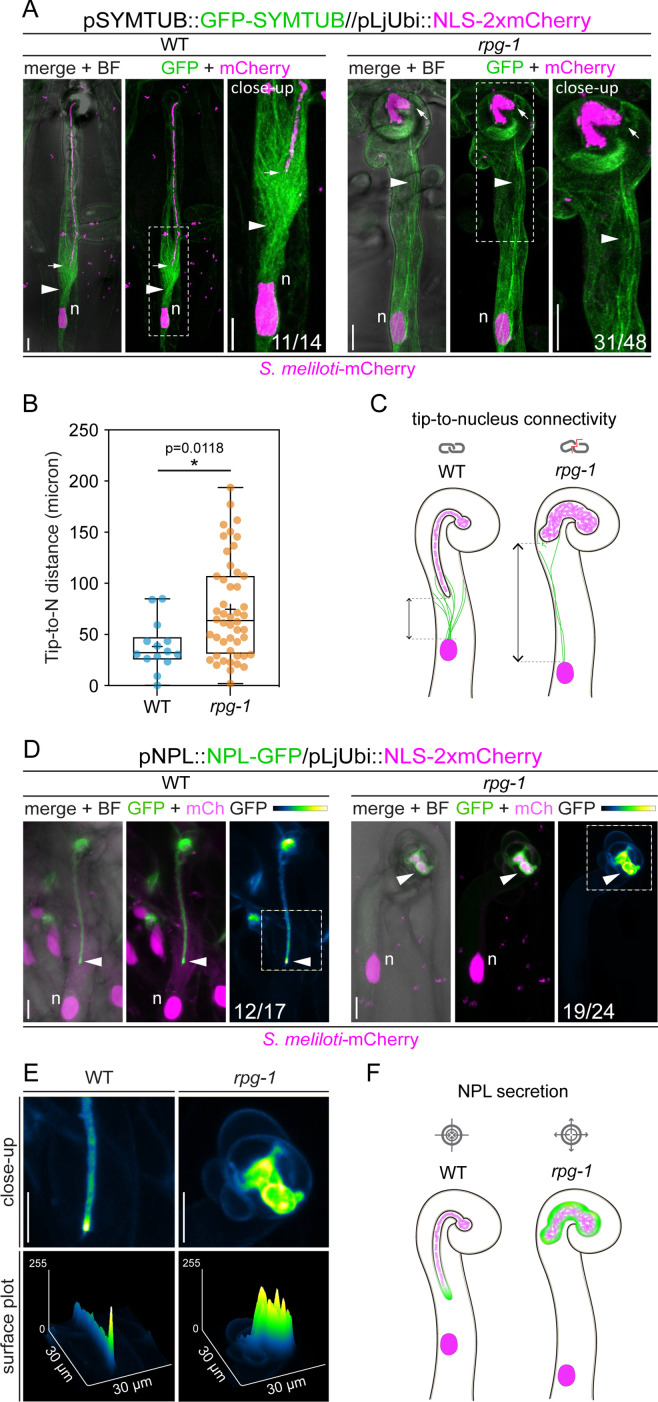
Figure 6. The tip-to-nucleus microtubule connectivity and polar secretion of Nodule pectate lyase (NPL) are affected in rpg-1.
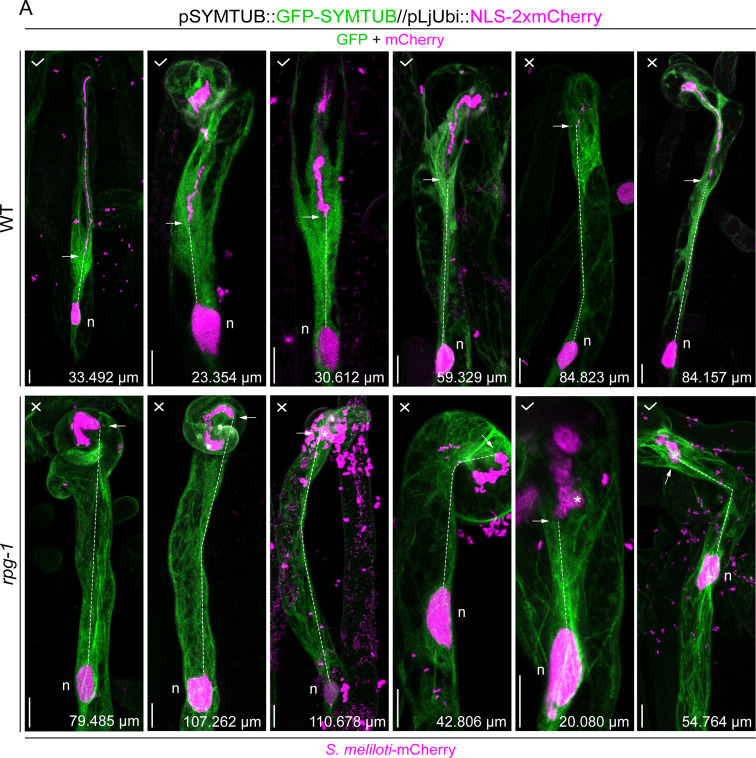
(A) In vivo imaging of microtubule patterning in root hairs hosting infection threads from wild-type (WT) and rpg-1 transgenic roots expressing GFP-SYMTUB at 4–8 dpi with S. meliloti-mCherry. The endoplasmic microtubule array (arrowhead) bridging the nucleus (n, magenta) and the infection thread (IT) tip (arrow) in WT root hairs appears dispersed and stretched in rpg-1 root hairs, where the IT tip does not follow the path of the migrating nucleus. A nuclear-localized tandem mCherry was used as a transformation marker. Images are merges of maximum intensity projections of GFP (green) and mCherry (magenta) channels after deconvolution of the corresponding stack series using the Huygens Professional software (Scientific Volume Imaging, The Netherlands). The bright field (BF) channel is overlaid with the merge. The dashed white line boxes indicate the region shown in the close-up. Numbers indicate frequencies of observation made on a total number of six (WT) and 14 (rpg-1) composite plants from two independent replicas. (B) Distance between the nucleus (N) and the IT tip measured in infected root hairs of WT and rpg-1 transgenic roots expressing GFP-SYMTUB. In the box plot, the top and bottom of each box represent the 75th and 25th percentiles, the middle horizontal bar indicates the median and the whiskers represent the range of minimum and maximum values. Crosses represent sample means. Asterisks indicate statistical significance based on a Mann-Whitney test with p-values <0.05 (*), <0.01 (**), <0.001 (***), and <0.0001 (****). n=14 (WT) and 48 (rpg-1) infected root hairs. (C) Schematic representation of infected root hairs showing endoplasmic microtubules (green curved lines) maintaining the nucleus (magenta oval) and the IT tip at a close distance (double-arrow line) in WT. Such tip-to-nucleus connectivity (gray icon) is altered (gray icon with red line) in rpg-1 infected root hairs, where microtubules appear rarefied and do not bridge the IT tip and the nucleus, being separated by an increased distance. Dashed light gray lines indicate the position of the nucleus and the tip. Bacteria within infection threads are colored in light magenta. (D) Live-cell confocal images showing focal accumulations of NPL-GFP at the IT tip (arrowhead) in WT root hairs in contrast to its unrestricted distribution in the apoplastic space surrounding ITs developed within root hairs of rpg-1 transgenic roots. A nuclear-localized tandem mCherry was used as a transformation marker. Images are maximum intensity projections. The GFP channel is shown in green when merged with the mCherry channel (magenta), or in Green Fire Blue when isolated, with yellow indicating the maximum intensity and blue a low level of fluorescence. The bright field (BF) channel is overlaid with the merge. Numbers indicate frequencies of observations made on a total number of 10 (WT) and nine (rpg-1) composite plants from two independent replicas. (E) Close-up and surface plot of the regions bounded by the dashed white line boxes in (D). The signal of NPL-GFP in WT ITs is described by a major intensity peak positioned on the advancing tip compared to multiple intensity peaks detected on ITs of rpg-1. (F) Schematic representation of infected root hairs showing the accumulation of NPL (green) restricted to the apoplastic space surrounding the tip of WT ITs in contrast to its accumulation in a broader apoplastic domain in IT of rpg-1, indicating that targeted secretion of this enzyme (gray icon with arrows pointing to center) is not correctly maintained in this mutant (gray icon with arrows pointing outside). Bacteria within infection threads are colored in light magenta. Scale bars = 10 µm.