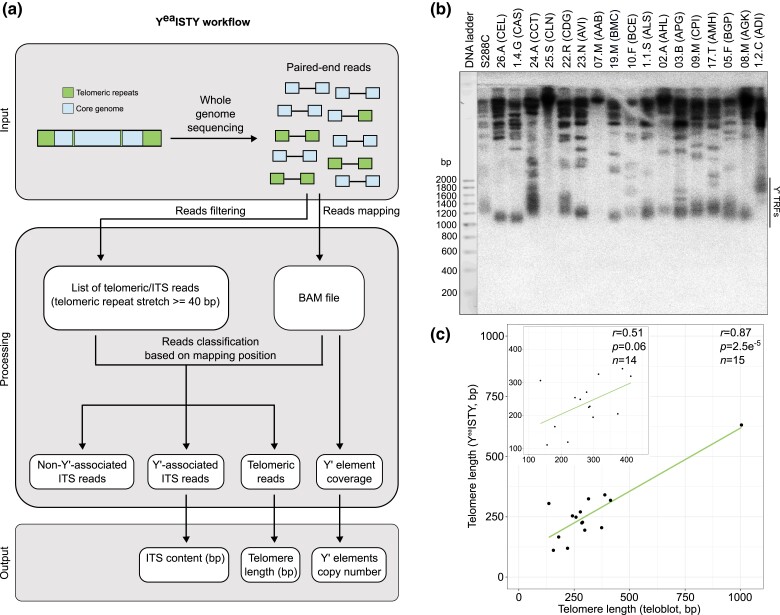
Fig. 1.
Overview of the YeaISTY workflow. a) Paired-end reads are scanned for telomeric repeats (either C{1,3}A or TG{1,3}). Reads containing a stretch of telomeric repeats longer or equal to 40 bp are retained. All the reads (retained and nonretained) are mapped to a modified reference genome. The mapping position of retained reads and their paired ones is used to classify them as ITS-derived or telomeric. The amount of ITS-derived and telomeric reads is then used to infer ITS content and telomere length, while coverage data are used to infer the copy number of Y′ elements. b) XhoI digestion of genomic DNA from representative strains of 17 S. cerevisiae lineages identified in Peter et al. (2018). Genomic DNA is probed with radioactively-labeled telomeric TG1–3 repeats. The black line denotes TRFs resulting from the digestion of a Y′ element. No TRFs were detected for the representatives of the Sake (25.S-CLN) and Mosaic beer (07.M-AAB) clades. The first lane shows a 200 bp DNA ladder used to derive the size of each TRF. c) Comparison of telomere length measured by teloblot and YeaISTY. Values in the plot are corrected for the 4-fold YeaISTY underestimation bias. Despite we estimated TL of S288C from sequencing data derived from Yue et al. (2017), we did not introduce this value in the plot as it derived from another sequencing batch and had a different underestimation rate. The Sake (CLN) and Mosaic beer (AAB) representatives were excluded from the comparison. The inset shows the correlation excluding the outlier with extremely long telomeres (1.2.C-ADI).