Fig. 3.
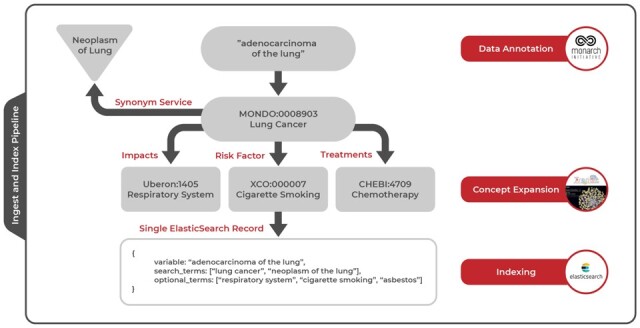
Detailed example of ingest and index pipeline. After ingesting a variable called ‘adenocarcinoma of the lung’ from study metadata, Dug uses NLP methods to annotate the variable with an ontology identifier for ‘Lung Cancer’ from the Mondo disease ontology. The resulting identifier is used to gather synonyms for lung cancer such as ‘Neoplasm of lung’ from an external API service. During concept expansion, Dug leverages TranQL to query knowledge graphs for other ontological concepts related to lung cancer through certain predicates; in the figure, we are looking for risk factors, treatments and anatomical entities impacted by lung cancer. During indexing, all terms discovered through annotation and concept expansion are combined with the original metadata into a single Elasticsearch record so that queries against any of these terms will yield the initial variable measuring ‘adenocarcinoma of the lung’
