Figure 1. The C-terminal helix of Ku80 can interact with DNA-PKcs in both cis and trans, but ablating the cis interaction promotes end joining.
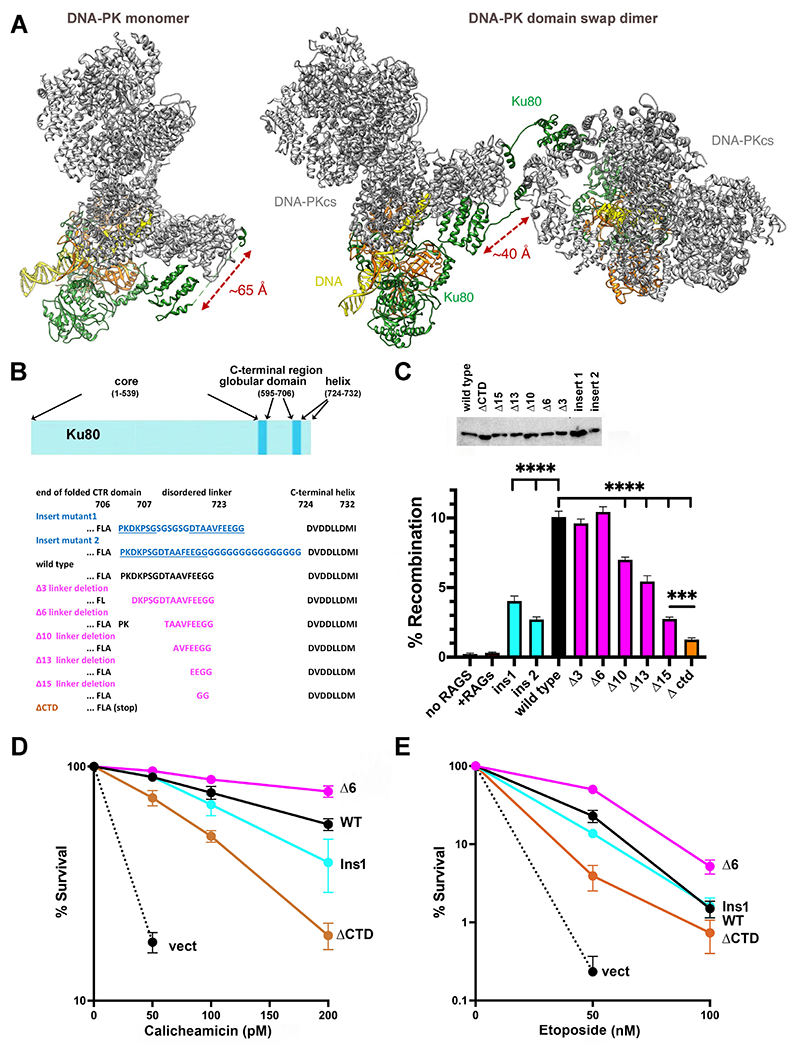
(A) Ribbon diagram of DNA-PK monomer and domain swap dimer. DNA-PKcs is shown in grey, Ku80 in green, Ku70 in orange and DNA in yellow. Diagram representing Ku80. A disordered linker (707-723) separates the C-terminal globular domain (595-706) from the C-terminal helix (724-732). (B) Amino acid sequence of the linker mutants. (C) The fluorescent substrate 290-Crimson/ZS was utilized to detect coding end joining of RAG-induced DSBs. Percent recombination of episomal fluorescent coding-end joining substrate in xrs6 cells transiently transfected with wild-type or mutant Ku80 expression constructs as indicated. Error bars indicate SEM from six independent experiments. ****, P<.0001; ***, P<.001 in two-way A NOVA with Holm-Sidak correction. Relative expression of each Ku80 mutant was assessed by immunoblotting. (D) + (E) xrs6 clonal transfectants expressing wild-type human Ku80 (WT), Δ6 linker shortening mutant (Δ6), insert 1 linker lengthening mutant (Ins1), Ctd linker/C-terminal+helix deletion mutant (ΔCtd), or empty vector (vect) were plated at cloning densities into complete medium with increasing doses of calicheamicin (D) or etoposide (E). Colonies were stained after eight days, and percent survival was calculated. Error bars represent the standard error of the means for six independent experiments.
