Figure 6. More disruptive mutants designed to disrupt the domain-swap and XLF-mediated DNA-PK dimers enhance their cellular phenotypes.
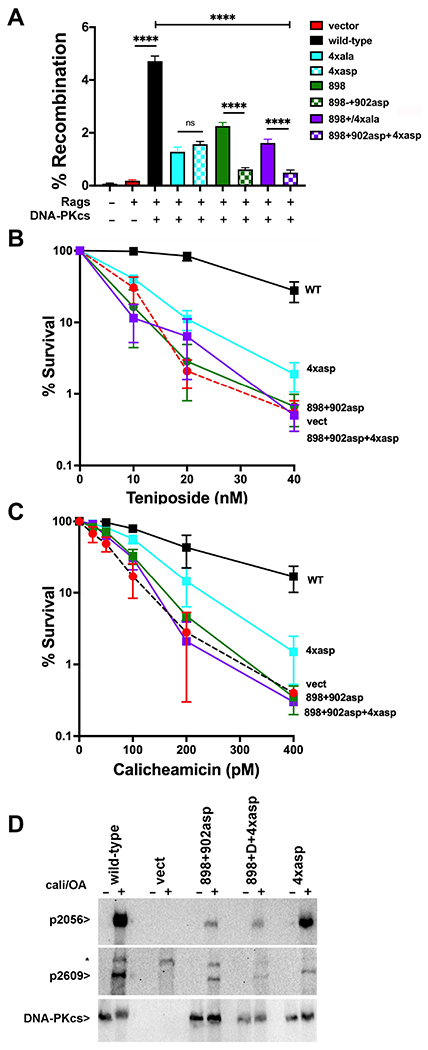
(A) (left) The fluorescent substrate 290-Crimson/ZS (coding joints) was utilized to detect coding end joining of RAG-induced DSBs in V3 cells expressing wild-type or mutant DNA-PKcs as depicted. Percent recombination represents %GFP/%Crimson. **P<0.01; ****P<0.0001; ns=not significant in two-way ANOVA with Holm-Sidak correction, (right) Immunoblot depicting relative DNA-PKcs expression in stable V3 clones as indicated. (B+C) V3 clonal transfectants expressing wild-type DNA-PKcs, vector control or DNA-PKcs mutants as indicated were plated at cloning densities into complete medium with increasing doses of calicheamicin (B) or teniposide (C). Colonies were stained after eight days, and percent survival was calculated. Error bars represent the standard error of the means for six independent experiments. (C) Immunoblot analyses using indicated antibodies of cell extracts from indicated V3 transfectants treated or not with 40nM calicheamicin and 1uM Okadaic acid for thirty minutes. * Indicates a non-specific band.
