Figure 4. SPOP W22R forms dramatically altered filaments.
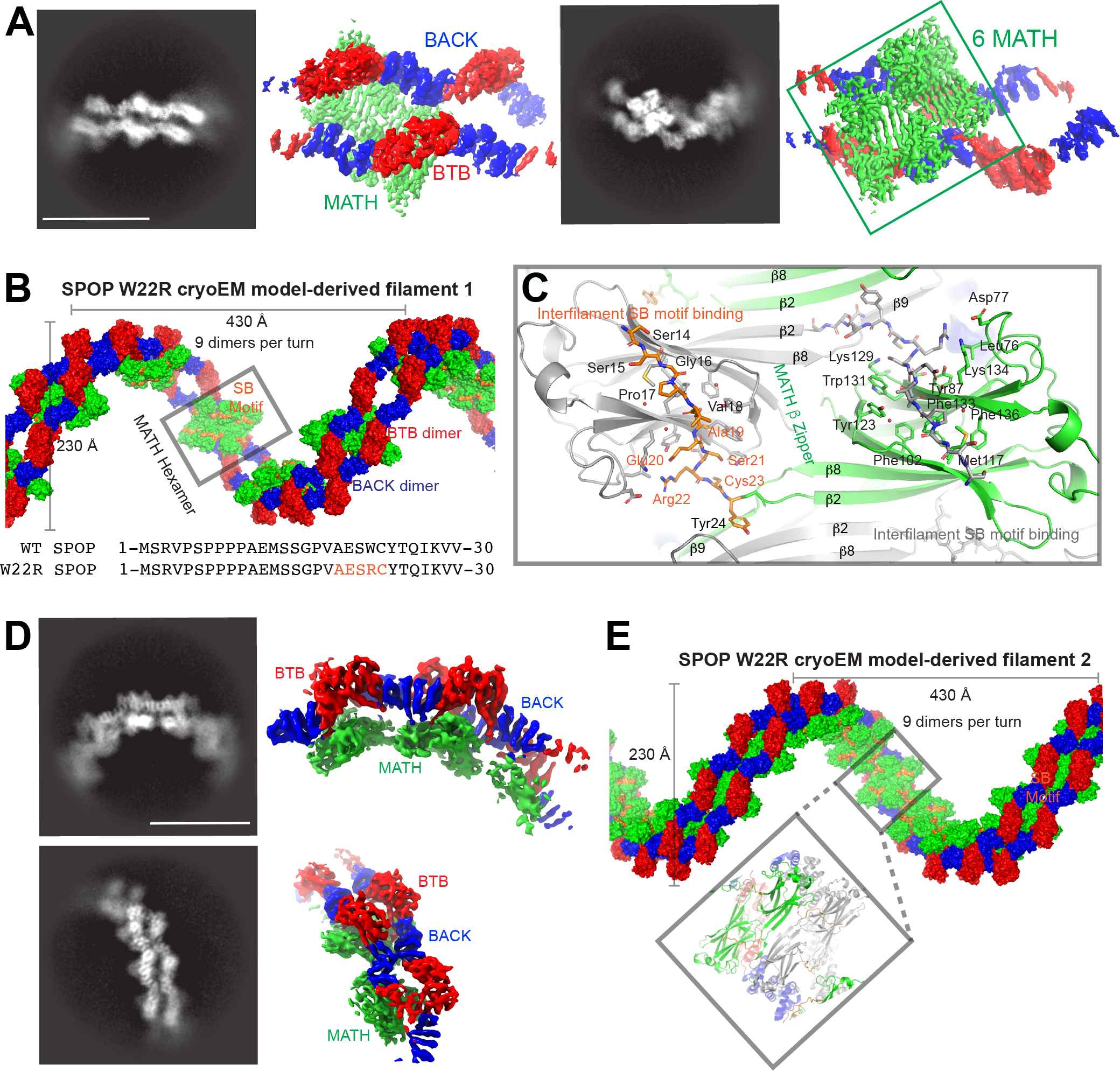
(A) Representative 2D classes (left) and sharpened cryo-EM maps in similar orientation (right) of SPOP W22R double filaments with MATH hexamer assemblies, i.e., population 1 (see also Figure S3).
(B) The cryo-EM model of population 1 of SPOP W22R can be used to construct a left-handed helical filament. Six ordered MATH domains (green surface, grey box) form a continuous assembly that link two individual SPOP filaments. The assembly is further held together by inter-monomer and inter-filament interactions of the substrate binding cleft in each MATH domain with a pseudo-SB motif sequence generated by mutation W22R (orange surface representation). Bottom, sequence alignment of N-terminal 30 amino acids of SPOP WT and W22R. Pseudo-SB motif in orange.
(C) Close up view of MATH domains. The β2 strands intercalate across the double filament in a zipper-like fashion (β-zipper) and link together two SPOP filaments. The R22-containing pseudo-SB motif binds to adjacent monomers and further links the two filaments together. MATH monomers from one filament are colored grey, whereas MATH monomers from the other filament are colored green; the N-terminus bound in the SB binding cleft is colored orange. See also Figure S6 for representations of the connectivity.
(D) Representative 2D classes (left) and sharpened cryo-EM maps in similar orientation (right) of SPOP W22R assemblies with MATH tetramer assemblies, i.e., population 2 (see also Figure S4).
(E) Population 2 of SPOP W22R particles have an alternative arrangement with a tetramer repeating unit. MATH tetramers link together two filaments. Helical parameters are similar to the population 1 of the SPOP W22R double filament.
