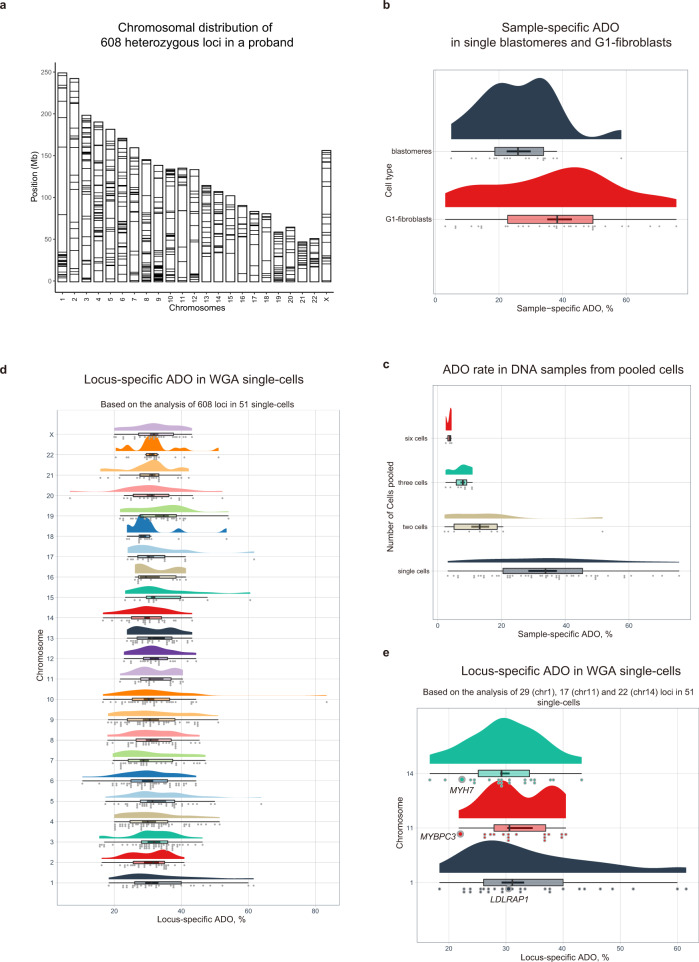
Fig. 2. Allelic dropouts due to whole-genome amplification biases limit the genotyping accuracy in human preimplantation embryos.
a Chromosomal distribution of 608 known heterozygous loci in female fibroblast donor used in the custom Ampliseq panel for investigation of ADO rates in single fibroblasts and blastomeres from SCNT embryos. b Distribution of sample-specific ADO rates observed in individual fibroblasts synchronized at G0–G1 phase of the cell cycle (n = 31) and blastomeres of cycling cleavage stage SCNT embryos (n = 18); n represents biologically independent number of samples collected over four independent experiments. c Distribution of ADO rates in WGA single-cell DNA (n = 51) and WGA DNA samples pooled from two cells (n = 9), three cells (n = 8) and six cells (n = 3); n represents biologically independent number of samples. d Chromosome- and locus-specific ADO rates for each of 608 targeted heterozygous loci. Based on the data from n = 51 single-cells examined over four independent experiments. e Frequency of ADO in three genomic regions closest to CRISPR/Cas9-targeted MYBPC3, MYH7, and LDLRAP1 loci. Based on the data from 51 single-cells examined over four independent experiments. For each box plot in (b–e): center line, median; box bounds, 25th and 75th percentiles; whiskers, minimum to maximum within 1.5 interquartile range; data points outside whiskers, outliers. Source Data are provided as Supplementary Data 5 and 6.