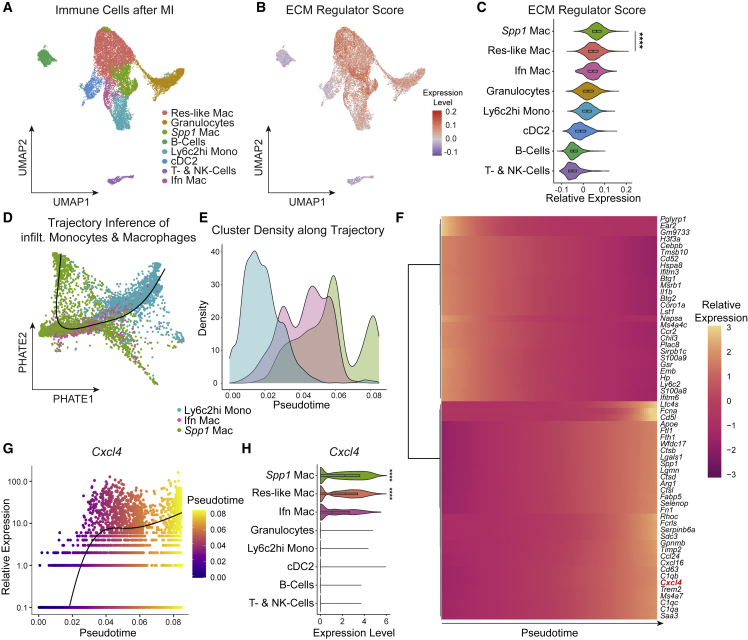
Figure 1.
ECM regulator scoring identifies profibrotic Spp1+ macrophages
(A) UMAP embedding of 17,690 Cd45+ immune cells from murine heart tissue at different timepoints after myocardial infarction from Forte et al.23 Labels refer to clusters. Res-like Mac: resident-like macrophages, Spp1 Mac: Spp1+ macrophages, Ly6c2hi Mono: Ly6c2 high monocytes, cDC2: conventional dendritic cells type 2, T- & NK-cells: T-cells and natural killer cells, Ifn Mac: Interferon-induced macrophages.
(B) Featureplot of ECM regulator score on the UMAP embedding shown in (A).
(C) ECM regulator score stratified by immune cell type.
(D) Fitted Slingshot pseudotime trajectory for infiltrating MPC on a PHATE dimensionality reduction.
(E) Line graph showing Cluster Density (in % of all cluster cells) of infiltrating MPC along pseudotime.
(F) Heatmap of top dynamically expressed genes along pseudotime.
(G) Expression of Cxcl4 along pseudotime. Each dot represents an individual pseudotime-ordered cell.
(H) Cxcl4 expression stratified by immune cell type.
For (C), a two-tailed unpaired t test (Spp1 Mac versus Res-like Mac) was computed. For (H) p values from MAST (Seurat, FindAllMarkers) are displayed. ∗∗∗∗p < 0.0001. See also Figures S1 and S2.