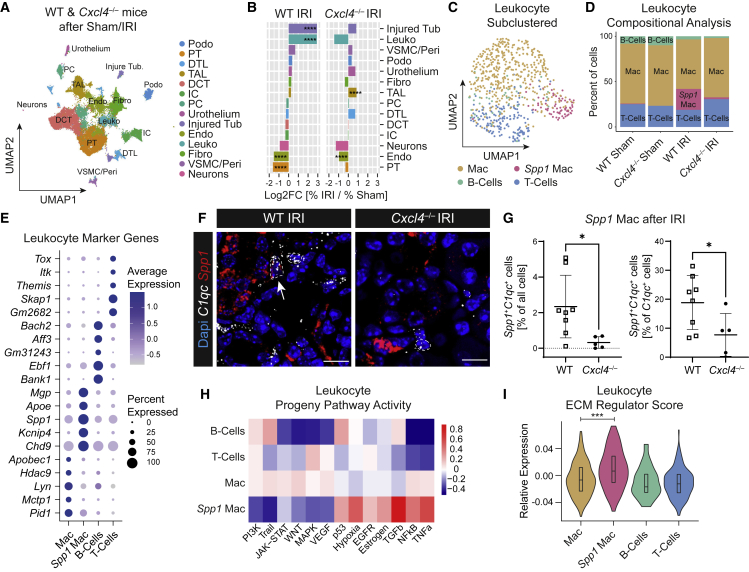
Figure 3.
Loss of Cxcl4 abrogates profibrotic Spp1+ macrophage expansion
(A) UMAP embedding of 66,235 nuclei isolated from kidneys of WT and Cxcl4−/− mice after sham or IRI surgery (n = 1 snRNA-seq library per condition pooled from n = 5 WT Sham, n = 5 WT IRI, n = 4 Cxcl4−/− Sham, and n = 4 Cxcl4−/− IRI mice). Labels refer to clusters. DCT, distal convoluted tubule; DTL, descending thin limb; Endo, endothelial cells; Fibro, fibroblasts; IC, intercalated cells; Injured Tub, injured tubular cells; Leuko, leukocytes; PC, principal cells; Podo, podocytes; PT, proximal tubule; TAL, thick ascending limb; Peri, pericytes; VSMC, vascular smooth muscle cells.
(B) Bar plot of cluster cell numbers in IRI versus sham kidneys for WT and Cxcl4−/− mice after normalization via Log2 transformation. Log2FC, log 2-Fold Change.
(C) UMAP of 489 sub-clustered single cell leukocytes from (A).
(D) Leukocyte sub-cluster composition (in % of all leukocytes) stratified by genotype and surgery.
(E) Dotplot of the top five specific genes for leukocyte clusters shown in (C).
(F) RNA-ISH staining for C1qc (white) and Spp1 (red) in WT and Cxcl4−/− kidneys after IRI. A white arrow marks a Spp1+C1qc+ cell (WT mice = 8, Cxcl4−/− mice = 5). Scale bar =10 μm.
(G) Quantification of C1qc+Spp1+ double-positive nuclei in percent of all nuclei and of C1qc+ nuclei. Data are shown as mean ± SD.
(H) PROGENy pathway analysis of snRNA leukocyte clusters.
(I) Leukocyte ECM regulator score stratified by leukocyte sub-cluster.
For (B) Fisher’s exact test was computed using false discovery rate correction for multiple testing. For (G) and (I) a two-tailed unpaired t test was performed. ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. See also Figures S3, S4, and S6.